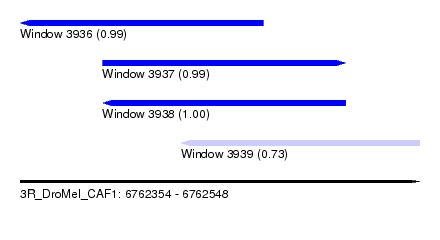
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,762,354 – 6,762,548 |
| Length | 194 |
| Max. P | 0.998881 |

| Location | 6,762,354 – 6,762,472 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -31.91 |
| Energy contribution | -32.11 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6762354 118 - 27905053 GAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACAUCAAUGUCAACAGGACAGCAG--CACACCGCCCCCAAAUGCCCUGUCCUCGCCGAAAUUCAUGGGACAACGUGGCCUGCA ......((....((((((((((((........(((((.......)))))..(((((((..(--((.............))).)))))))............))))....)))))))))). ( -35.42) >DroSec_CAF1 26595 120 - 1 GAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACAUCAAUGUCAACAGGACAGCAGGGCACACCGCCCCCAAAUGCCCUGUCCUCGCCGAAAUUCAUGGGGCAACGUGGCCUGCA (.(((((((....))))).)).).........(((((.......)))))........(((((.(((...(((((...(((.....((......))....))))))))...))).))))). ( -44.00) >DroSim_CAF1 30935 120 - 1 GAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACAUCAAUGUCAACAGGACAGCAGCGCACACCGCCCCCAAAUGCCCUGUCCUCGCCGAAAUUCAUGGGGCAACGUGGCCUGCA (.(((((((....))))).)).).........(((((.......)))))........((((.((.(((.(((((...(((.....((......))....))))))))...))))))))). ( -40.30) >DroEre_CAF1 11043 118 - 1 GAGCUGGCCGAAAGGCCACGCCCAACGCAACAUGACAACAUCAAUGUCAACAGGACAGCAG--CACACCGCCCCCAAAUGCCCUGUCCUCUCCGAAAUUCAUGGGACAACGUGGCCUGCA (.(((((((....))))).)).)...(((...(((((.......)))))..(((((((..(--((.............))).))))))).........(((((......)))))..))). ( -35.82) >DroYak_CAF1 23202 118 - 1 GAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACAUCAAUGUCAACAGGACAGCAG--CACACCGCCCCCAAAUGCCCUGUCCUCCACGAAAUUCAUGGCACAACGUGGCCUGCA (.(((((((....))))).)).).........(((((.......)))))..(((((((..(--((.............))).)))))))(((((...............)))))...... ( -33.98) >consensus GAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACAUCAAUGUCAACAGGACAGCAG__CACACCGCCCCCAAAUGCCCUGUCCUCGCCGAAAUUCAUGGGACAACGUGGCCUGCA ......((....((((((((((((........(((((.......)))))..(((((((((..................)))..))))))............))))....)))))))))). (-31.91 = -32.11 + 0.20)


| Location | 6,762,394 – 6,762,512 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -44.34 |
| Consensus MFE | -39.36 |
| Energy contribution | -39.96 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6762394 118 + 27905053 CAUUUGGGGGCGGUGUG--CUGCUGUCCUGUUGACAUUGAUGUUGUCAUGUUGUGUUGGGCGUGGCCUUUCGGCCAGCUCGGCAAAUACCUGUUUAUGUUAUGCCUGGCACAUCAGAGCA ......((((((((...--..))))))))(((((((.......)))).((.(((((..(((((((((....))))(((..((((......))))...))))))))..))))).)).))). ( -43.90) >DroSec_CAF1 26635 120 + 1 CAUUUGGGGGCGGUGUGCCCUGCUGUCCUGUUGACAUUGAUGUUGUCAUGUUGUGUUGGGCGUGGCCUUUCGGCCAGCUCGGCAAAUACCUGUUUAUGUUAUGCCUGCCACAUCAGGGCA ......(((((.....)))))...((((((.(((((.......)))))...((((..((((.(((((....)))))))))((((((((........)))).))))...)))).)))))). ( -44.90) >DroSim_CAF1 30975 112 + 1 CAUUUGGGGGCGGUGUGCGCUGCUGUCCUGUUGACAUUGAUGUUGUCAUGUUGUGUUGGGCGUGGCCUUUCGGCCAGCUCGGCAAAUACCUGUUUAUGUUAUGCCUGGCACA-------- ......((((((((.......))))))))..(((((.......)))))...(((((..(((((((((....))))(((..((((......))))...))))))))..)))))-------- ( -41.50) >DroEre_CAF1 11083 118 + 1 CAUUUGGGGGCGGUGUG--CUGCUGUCCUGUUGACAUUGAUGUUGUCAUGUUGCGUUGGGCGUGGCCUUUCGGCCAGCUCGGCAAAUACCUGUUUAUGUUAUGCCUGGCACAUCAGGGCG ......((((((((...--..))))))))..(((((.......)))))....(((((((((.(((((....)))))))))))).....((((....(((((....)))))...)))))). ( -44.60) >DroYak_CAF1 23242 118 + 1 CAUUUGGGGGCGGUGUG--CUGCUGUCCUGUUGACAUUGAUGUUGUCAUGUUGUGUUGGGCGUGGCCUUUCGGCCAGCUCGGCAAAUACCUGUUUAUGUUAUGCCUGGCACAUCAGGGCA ........(((((....--)))))((((((.(((((.......)))))...(((((..(((((((((....))))(((..((((......))))...))))))))..))))).)))))). ( -46.80) >consensus CAUUUGGGGGCGGUGUG__CUGCUGUCCUGUUGACAUUGAUGUUGUCAUGUUGUGUUGGGCGUGGCCUUUCGGCCAGCUCGGCAAAUACCUGUUUAUGUUAUGCCUGGCACAUCAGGGCA ......((((((((.......))))))))..(((((.......)))))((.((((..((((((((((....))))(((..((((......))))...)))))))))..)))).))..... (-39.36 = -39.96 + 0.60)


| Location | 6,762,394 – 6,762,512 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -35.86 |
| Consensus MFE | -30.46 |
| Energy contribution | -30.98 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6762394 118 - 27905053 UGCUCUGAUGUGCCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACAUCAAUGUCAACAGGACAGCAG--CACACCGCCCCCAAAUG (((((((.((((...((((..((........))...))))(.(((((((....))))).)).)..))))...(((((.......)))))......))).))--))............... ( -35.30) >DroSec_CAF1 26635 120 - 1 UGCCCUGAUGUGGCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACAUCAAUGUCAACAGGACAGCAGGGCACACCGCCCCCAAAUG (((((((.((((...((((..((........))...))))(.(((((((....))))).)).)..))))...(((((.......))))).))))...)))((((.....))))....... ( -42.80) >DroSim_CAF1 30975 112 - 1 --------UGUGCCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACAUCAAUGUCAACAGGACAGCAGCGCACACCGCCCCCAAAUG --------(((((...((....(((.....(((....)))(.(((((((....))))).)).)........)))..........((((.....))))))...)))))............. ( -31.10) >DroEre_CAF1 11083 118 - 1 CGCCCUGAUGUGCCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACGCAACAUGACAACAUCAAUGUCAACAGGACAGCAG--CACACCGCCCCCAAAUG .((((((((((((..((((..((........))...))))(.(((((((....))))).)).)...)).))))((((.......))))..))))...)).(--(.....))......... ( -34.80) >DroYak_CAF1 23242 118 - 1 UGCCCUGAUGUGCCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACAUCAAUGUCAACAGGACAGCAG--CACACCGCCCCCAAAUG (((((((.((((...((((..((........))...))))(.(((((((....))))).)).)..))))...(((((.......))))).))))...)))(--(.....))......... ( -35.30) >consensus UGCCCUGAUGUGCCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACAUCAAUGUCAACAGGACAGCAG__CACACCGCCCCCAAAUG ...((((.((((...((((..((........))...))))(.(((((((....))))).)).)..))))...(((((.......))))).)))).......................... (-30.46 = -30.98 + 0.52)



| Location | 6,762,432 – 6,762,548 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.24 |
| Mean single sequence MFE | -36.19 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.92 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6762432 116 - 27905053 AUACUGGCCA----ACAACGGGGGAAUGGUGUAGGGAUGAUGCUCUGAUGUGCCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACA ...(((....----....))).(..(((((((.(((.((..((((((.(((((...))))).))......(((....)))))))(((((....)))))))))).))))..)))..).... ( -38.00) >DroSec_CAF1 26675 116 - 1 CUACUGGGCA----ACAACGGGGGAAUGGGGUAGGGAUGAUGCCCUGAUGUGGCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACA ....(((((.----(((.(((((..((.(........).)).))))).)))(((.((((..((........))...))))..)))((((....))))..)))))................ ( -38.50) >DroSim_CAF1 31015 105 - 1 AUACUGGGCA----ACAACGGGGGAAUGGGGUAGGGA-----------UGUGCCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACA ......(...----.)...(..(..(((..((.(((.-----------(((((..((((..((........))...))))..)).((((....))))))))))...))..)))..)..). ( -33.40) >DroEre_CAF1 11121 108 - 1 ACACUGGGCA----ACAACG--------UGGUAAGGAGGACGCCCUGAUGUGCCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACGCAACAUGACAACA ....(((((.----......--------(((((...(((....)))....)))))((((..((........))...))))..(.(((((....)))))))))))................ ( -34.90) >DroYak_CAF1 23280 116 - 1 GCAGUGGACAGUGGACAGGG----UAAGGAGUAAGGAGGAUGCCCUGAUGUGCCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACA ...((.....(((..(((((----((..............))))))).((((...((((..((........))...))))(.(((((((....))))).)).)..)))).)))....)). ( -36.14) >consensus AUACUGGGCA____ACAACGGGGGAAUGGGGUAGGGAGGAUGCCCUGAUGUGCCAGGCAUAACAUAAACAGGUAUUUGCCGAGCUGGCCGAAAGGCCACGCCCAACACAACAUGACAACA ....(((((..........................................((..((((..((........))...))))..))(((((....))))).)))))................ (-25.32 = -25.92 + 0.60)

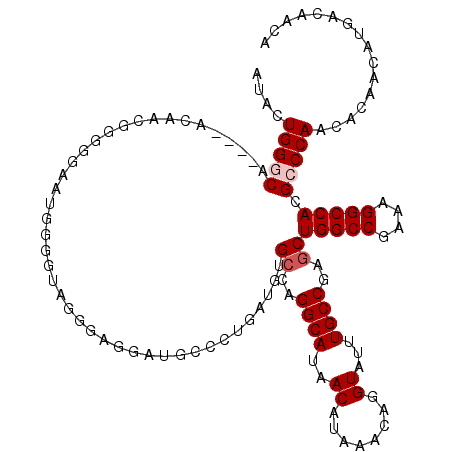
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:26 2006