
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,761,664 – 6,761,844 |
| Length | 180 |
| Max. P | 0.999206 |

| Location | 6,761,664 – 6,761,784 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -30.72 |
| Energy contribution | -31.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
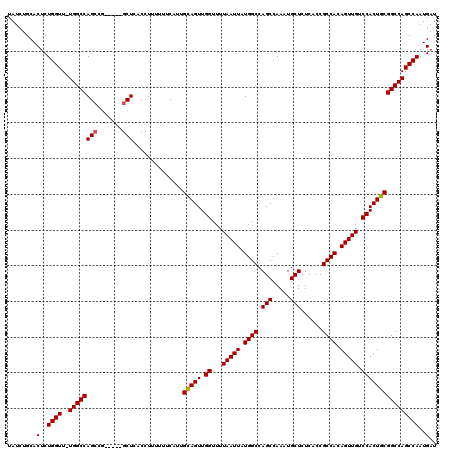
>3R_DroMel_CAF1 6761664 120 + 27905053 CAUCCCUGCACGGCUGCUGCCAACAUCAUAUCCAAUUAACAUCAUUGGCUGGCCGCAGUGGACAACUGUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUGCAAUGAAAAA ..((..((((.(((....)))........................((((..((((((((.....)))))....((....))....)))..))))..............))))..)).... ( -33.50) >DroSec_CAF1 25877 120 + 1 CAUCCCAGCACGACUGCUGCCAACAUCAUAUCCAAUUAACAUCAUUGGCUGGCCGCAGUGGACAACUGUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUACAAUGAAAAA ....(((((.(((.(((((((..........(((((.......)))))...((((((((.....)))))))))))...)))).))))))))............................. ( -35.30) >DroSim_CAF1 30262 120 + 1 CAUCCCAGCACGACUGCUGCCAACAUCAUAUCCAAUUAACAUCAUUGGCUGGCCGCAGUGGACAACUGUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUGCAAUGAAAAA ....(((((.(((.(((((((..........(((((.......)))))...((((((((.....)))))))))))...)))).))))))))............................. ( -35.30) >DroEre_CAF1 10346 120 + 1 AGUCCCAGCAUGGCGGAUGCCAACAUCAUAUCCAAUUAACAUCAUUGGCUGGCCGCAGUGGACAACUGUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUGCAAUGGAAAA ....(((((....(((((((...((((....(((((.......)))))...((((((((.....))))))))))))...))))))))))))............(((.......))).... ( -36.20) >DroYak_CAF1 22507 119 + 1 CAUCCCAGCACGACUGCUGCCAACAUCAUAUCCAAUUAACAUCAUUGGCUGGCCGCAGUGGACAACUAUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUGCAAUG-AAAA ....(((((.(((.((((.............(((((.......)))))((.(((((.((((....)))).))))).)))))).))))))))........................-.... ( -32.10) >consensus CAUCCCAGCACGACUGCUGCCAACAUCAUAUCCAAUUAACAUCAUUGGCUGGCCGCAGUGGACAACUGUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUGCAAUGAAAAA ....(((((.(((.(((((((..........(((((.......)))))...((((((((.....)))))))))))...)))).))))))))............................. (-30.72 = -31.12 + 0.40)

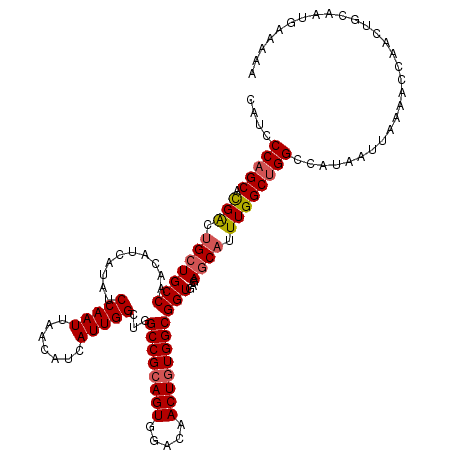

| Location | 6,761,704 – 6,761,818 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.76 |
| Mean single sequence MFE | -38.44 |
| Consensus MFE | -33.74 |
| Energy contribution | -33.74 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6761704 114 + 27905053 AUCAUUGGCUGGCCGCAGUGGACAACUGUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUGCAAUGAAAAAAGGUGAGC-----UGGCUGGCCA-AACCAGAGUGCAGAUA (((.((.((((.(((((((.....))))))))))).)).(((((((((..((((........(((................)))....-----))))..))))-)......)))).))). ( -38.69) >DroSec_CAF1 25917 114 + 1 AUCAUUGGCUGGCCGCAGUGGACAACUGUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUACAAUGAAAAAAGGAGAGC-----CGGCUGGCCA-AACCAGAGUGCAGAUA (((.((.((((.(((((((.....))))))))))).)).(((((((((..(((..............(......)......((....)-----))))..))))-)......)))).))). ( -38.10) >DroSim_CAF1 30302 114 + 1 AUCAUUGGCUGGCCGCAGUGGACAACUGUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUGCAAUGAAAAAAGGUGAGC-----CGGCUGGCCA-AACCAGAGUGCAGAUA (((.((.((((.(((((((.....))))))))))).)).(((((((((..(((..............(......)......((....)-----))))..))))-)......)))).))). ( -39.00) >DroEre_CAF1 10386 119 + 1 AUCAUUGGCUGGCCGCAGUGGACAACUGUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUGCAAUGGAAAAAAGUGAGUGUUCACCGCUGGCCA-AACCAGAGUGCAGAUA (((.((.((((.(((((((.....))))))))))).)).((((((((.(((((..........(((.......))).....((((.((....)))))))))))-..))))).))).))). ( -40.90) >DroYak_CAF1 22547 114 + 1 AUCAUUGGCUGGCCGCAGUGGACAACUAUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUGCAAUG-AAAAAAGUGAGC-----CGGCUGGCCAAAACCAGAGUGCAGAUA .....((((..(((((.((((....)))).)).((....))....)))..)))).............(((((...-.......((.((-----(....))))).........)))))... ( -35.51) >consensus AUCAUUGGCUGGCCGCAGUGGACAACUGUGGCGGUGAGAGCAUUUGGCUGGCCAUAAUUAAAACCAACUGCAAUGAAAAAAGGUGAGC_____CGGCUGGCCA_AACCAGAGUGCAGAUA (((.((.((((.(((((((.....))))))))))).)).((((((((.(((((.............(((............))).(((.......))))))))...))))).))).))). (-33.74 = -33.74 + -0.00)

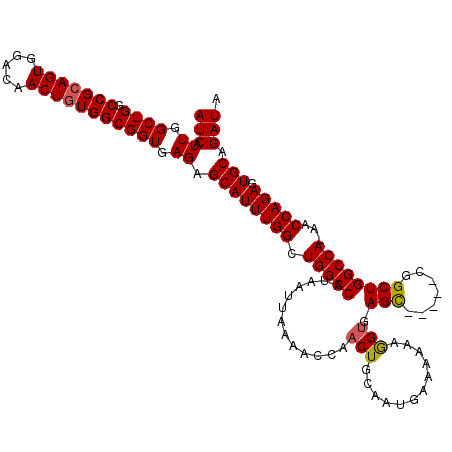

| Location | 6,761,704 – 6,761,818 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.76 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6761704 114 - 27905053 UAUCUGCACUCUGGUU-UGGCCAGCCA-----GCUCACCUUUUUUCAUUGCAGUUGGUUUUAAUUAUGGCCAGCCAAAUGCUCUCACCGCCACAGUUGUCCACUGCGGCCAGCCAAUGAU .....(((...(((((-.(((((((((-----(((((...........)).)))))))........))))))))))..))).......(((.((((.....)))).)))........... ( -32.70) >DroSec_CAF1 25917 114 - 1 UAUCUGCACUCUGGUU-UGGCCAGCCG-----GCUCUCCUUUUUUCAUUGUAGUUGGUUUUAAUUAUGGCCAGCCAAAUGCUCUCACCGCCACAGUUGUCCACUGCGGCCAGCCAAUGAU .....(((...(((((-.(((((((((-----((((.............).)))))))........))))))))))..))).......(((.((((.....)))).)))........... ( -31.42) >DroSim_CAF1 30302 114 - 1 UAUCUGCACUCUGGUU-UGGCCAGCCG-----GCUCACCUUUUUUCAUUGCAGUUGGUUUUAAUUAUGGCCAGCCAAAUGCUCUCACCGCCACAGUUGUCCACUGCGGCCAGCCAAUGAU ........(..(((((-.((((....(-----(....))..........(((((.((...(((((.((((.(((.....)))......)))).))))).))))))))))))))))..).. ( -32.50) >DroEre_CAF1 10386 119 - 1 UAUCUGCACUCUGGUU-UGGCCAGCGGUGAACACUCACUUUUUUCCAUUGCAGUUGGUUUUAAUUAUGGCCAGCCAAAUGCUCUCACCGCCACAGUUGUCCACUGCGGCCAGCCAAUGAU ........(..(((((-.((((...(((((....)))))..........(((((.((...(((((.((((.(((.....)))......)))).))))).))))))))))))))))..).. ( -35.80) >DroYak_CAF1 22547 114 - 1 UAUCUGCACUCUGGUUUUGGCCAGCCG-----GCUCACUUUUUU-CAUUGCAGUUGGUUUUAAUUAUGGCCAGCCAAAUGCUCUCACCGCCAUAGUUGUCCACUGCGGCCAGCCAAUGAU .....((...(((((....)))))...-----)).........(-(((((((((.((...((((((((((.(((.....)))......)))))))))).)))))))((....))))))). ( -35.00) >consensus UAUCUGCACUCUGGUU_UGGCCAGCCG_____GCUCACCUUUUUUCAUUGCAGUUGGUUUUAAUUAUGGCCAGCCAAAUGCUCUCACCGCCACAGUUGUCCACUGCGGCCAGCCAAUGAU ........(..((((..((((((((.......)))..............(((((.((...(((((.((((.(((.....)))......)))).))))).))))))))))))))))..).. (-30.70 = -30.74 + 0.04)


| Location | 6,761,744 – 6,761,844 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.05 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6761744 100 - 27905053 ---ACU-----------GGCAAGGACUGCAGCGGCAUUUGUAUCUGCACUCUGGUU-UGGCCAGCCA-----GCUCACCUUUUUUCAUUGCAGUUGGUUUUAAUUAUGGCCAGCCAAAUG ---..(-----------(((..(((.(((((..((....))..))))).)))....-((((((((((-----(((((...........)).)))))))........)))))))))).... ( -32.70) >DroSec_CAF1 25957 100 - 1 ---ACU-----------GGCAAGGACUGCAGCGGCAUUUGUAUCUGCACUCUGGUU-UGGCCAGCCG-----GCUCUCCUUUUUUCAUUGUAGUUGGUUUUAAUUAUGGCCAGCCAAAUG ---..(-----------(((..(((.(((((..((....))..))))).)))....-((((((((((-----((((.............).)))))))........)))))))))).... ( -31.42) >DroSim_CAF1 30342 100 - 1 ---ACU-----------GGCAAGGACUGCAGCGGCAUUUGUAUCUGCACUCUGGUU-UGGCCAGCCG-----GCUCACCUUUUUUCAUUGCAGUUGGUUUUAAUUAUGGCCAGCCAAAUG ---..(-----------(((..(((.(((((..((....))..))))).)))....-((((((((((-----(((((...........)).)))))))........)))))))))).... ( -31.90) >DroEre_CAF1 10426 108 - 1 GACACU-----------GGGCAGUACUGCAGCGGCAUUUGUAUCUGCACUCUGGUU-UGGCCAGCGGUGAACACUCACUUUUUUCCAUUGCAGUUGGUUUUAAUUAUGGCCAGCCAAAUG ......-----------.(((...((((((((((.((....)))))).(.((((..-...)))).)((((....))))..........))))))(((((........))))))))..... ( -29.40) >DroYak_CAF1 22587 114 - 1 GACGCAGGAUGCAGGAUGGCAAGUACUGCAGCGGCAUUUGUAUCUGCACUCUGGUUUUGGCCAGCCG-----GCUCACUUUUUU-CAUUGCAGUUGGUUUUAAUUAUGGCCAGCCAAAUG (((.(((..((((((...((((((.(((...))).)))))).))))))..))))))(((((((((((-----(((((.......-...)).)))))))........)))))))....... ( -36.10) >consensus ___ACU___________GGCAAGGACUGCAGCGGCAUUUGUAUCUGCACUCUGGUU_UGGCCAGCCG_____GCUCACCUUUUUUCAUUGCAGUUGGUUUUAAUUAUGGCCAGCCAAAUG .................(((....((((((((((.((....))))))...(((((....)))))........................))))))(((((........))))))))..... (-25.12 = -25.16 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:22 2006