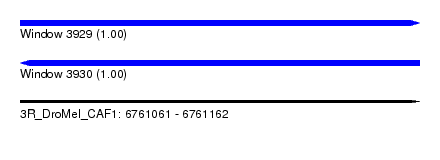
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,761,061 – 6,761,162 |
| Length | 101 |
| Max. P | 0.998369 |

| Location | 6,761,061 – 6,761,162 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -17.39 |
| Energy contribution | -20.31 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 0.92 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
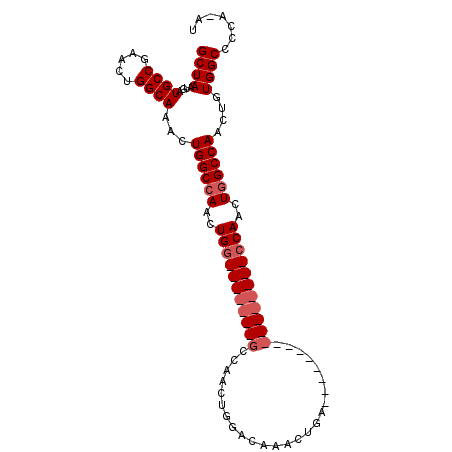
>3R_DroMel_CAF1 6761061 101 + 27905053 GCUAAUGAUGCCGAACUGGCAAACUGGCCAACUGGCCAACUGGCCAACUGGACAAACUGACCAACUGACCAACUGGCCAACUGGCCAACUGUGGCCCCA-AU ........((((.....))))....(((((..((((((..((((((..(((.((...........)).)))..))))))..))))))....)))))...-.. ( -38.50) >DroSim_CAF1 29681 77 + 1 GCUAAUGAUGCCGAACUGGCAAACUGGCCAACUG--------GCCAACUGGACAAACUGA----------------CCAACUGGCCAACUGUGGCCCCA-AU ........((((.....))))....(((((..((--------((((..(((.(.....).----------------)))..))))))....)))))...-.. ( -27.00) >DroEre_CAF1 9774 78 + 1 GCUAAUGAUGCCGAACUGGCAAACUGGCAAACUGG--------CCAACUCGACAAACUGG----------------CCAACUGGCCAACUGUGGCCCCAAAA ((((....((((.....))))...))))....(((--------(((...........)))----------------)))...(((((....)))))...... ( -24.50) >consensus GCUAAUGAUGCCGAACUGGCAAACUGGCCAACUGG_______GCCAACUGGACAAACUGA________________CCAACUGGCCAACUGUGGCCCCA_AU ((((....((((.....))))...((((((..(((((((((((.........................)))))))))))..))))))....))))....... (-17.39 = -20.31 + 2.92)

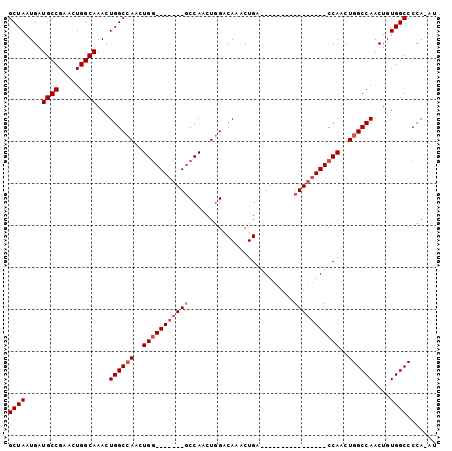
| Location | 6,761,061 – 6,761,162 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -23.85 |
| Energy contribution | -24.77 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
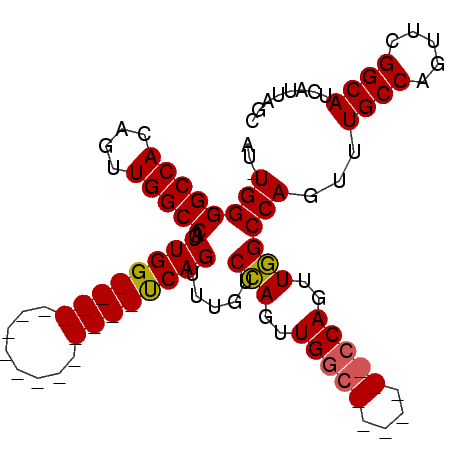
>3R_DroMel_CAF1 6761061 101 - 27905053 AU-UGGGGCCACAGUUGGCCAGUUGGCCAGUUGGUCAGUUGGUCAGUUUGUCCAGUUGGCCAGUUGGCCAGUUGGCCAGUUUGCCAGUUCGGCAUCAUUAGC ((-((......))))((((((..((((((..((((((..(((.(.....).)))..))))))..))))))..))))))...((((.....))))........ ( -48.70) >DroSim_CAF1 29681 77 - 1 AU-UGGGGCCACAGUUGGCCAGUUGG----------------UCAGUUUGUCCAGUUGGC--------CAGUUGGCCAGUUUGCCAGUUCGGCAUCAUUAGC ..-...(((((...(((((((..(((----------------.(.....).)))..))))--------))).)))))....((((.....))))........ ( -31.90) >DroEre_CAF1 9774 78 - 1 UUUUGGGGCCACAGUUGGCCAGUUGG----------------CCAGUUUGUCGAGUUGG--------CCAGUUUGCCAGUUUGCCAGUUCGGCAUCAUUAGC ..(((((((((....)))))..((((----------------(((..((...))..)))--------))))....))))..((((.....))))........ ( -26.90) >consensus AU_UGGGGCCACAGUUGGCCAGUUGG________________UCAGUUUGUCCAGUUGGC_______CCAGUUGGCCAGUUUGCCAGUUCGGCAUCAUUAGC ...((((((((....)))))..((((((((........)))))))).....(((..(((((....)))))..))))))...((((.....))))........ (-23.85 = -24.77 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:17 2006