
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,760,588 – 6,760,726 |
| Length | 138 |
| Max. P | 0.997659 |

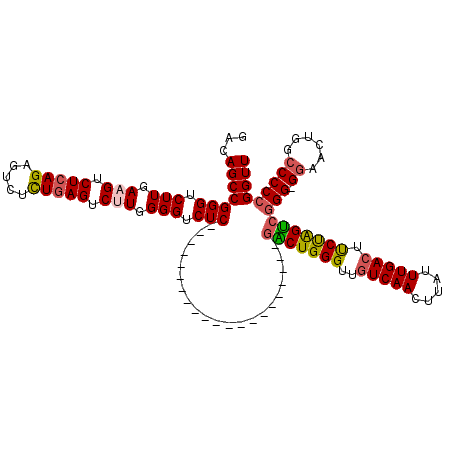
| Location | 6,760,588 – 6,760,686 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.34 |
| Mean single sequence MFE | -49.30 |
| Consensus MFE | -35.12 |
| Energy contribution | -35.40 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6760588 98 + 27905053 GACAGCCGGGUCUUGAAGUCUCAGAGUUUCUGAGUCUUGGGGUCUC---------------------GACUGGGUUGUCAACUUAUUUGACUCCUAGUCGGG-GGAACUGGCCCCCGGUU ...(((((((.(((.(((.(((((.....))))).))).))).)))---------------------(((((((..(((((.....))))).)))))))(((-((......))))))))) ( -48.60) >DroSec_CAF1 24831 98 + 1 GACAGCCGGGUCUUGAAGUCUCAGAGUCUUUGAGUCUUGGGGUCUC---------------------GACUGGGUUGUCAACUUAUUUGACUUCUAGUCGGG-GGAACUGGCCCCCGGUU ...(((((((.(((.(((.((((((...)))))).))).))).)))---------------------(((((((..(((((.....))))).)))))))(((-((......))))))))) ( -43.10) >DroSim_CAF1 29205 98 + 1 GACAGCCGGGUCUUGAAGUCUCAGAGUCUCUGAGUCUUGGGGUCUC---------------------GACUGGGUUGUCAACUUAUUUGACUUCUAGUCGGG-GGAACUGGCCCCCGGUU ...(((((((.(((.(((.(((((.....))))).))).))).)))---------------------(((((((..(((((.....))))).)))))))(((-((......))))))))) ( -46.00) >DroEre_CAF1 9291 106 + 1 GACAGCCGGGUCUUGAAGUCUCAGAGUCUCUGAGUCCUGGGGUCUCGGG--------------GCCCGGCUGGGUUGUCAACUUAUUUGAGUUCCAGUCGGGGGGAACUGGCCCCCGGUU ..((((((((((((((..((((((.(.(.....).)))))))..)))))--------------)))))))))((...((((.....))))...))(..((((((.......))))))..) ( -53.50) >DroYak_CAF1 21385 120 + 1 GACAGCCGGGUCUUGAAGUCUCAGAGUAUCUUAGUCCUGGGGUCUCGGGACUCGGUACUCGGGACUCGGCUGGGUUGUCAACUUAUUUGACUCCUGGCGAGGGGAAAGUGGCCCCCGGUU ..((((((((((((((....)).(((((((..((((((((....)))))))).)))))))))))))))))))((..(((((.....))))).))...((.((((.......))))))... ( -55.30) >consensus GACAGCCGGGUCUUGAAGUCUCAGAGUCUCUGAGUCUUGGGGUCUC_____________________GACUGGGUUGUCAACUUAUUUGACUUCUAGUCGGG_GGAACUGGCCCCCGGUU ...(((((((.(((.(((.(((((.....))))).))).))).))).....................(((((((..(((((.....))))).)))))))(((.((......))))))))) (-35.12 = -35.40 + 0.28)


| Location | 6,760,588 – 6,760,686 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.34 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.74 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6760588 98 - 27905053 AACCGGGGGCCAGUUCC-CCCGACUAGGAGUCAAAUAAGUUGACAACCCAGUC---------------------GAGACCCCAAGACUCAGAAACUCUGAGACUUCAAGACCCGGCUGUC ...((((((......))-)))).......(((((.....)))))....(((((---------------------(.(.....(((.(((((.....))))).))).....).)))))).. ( -31.70) >DroSec_CAF1 24831 98 - 1 AACCGGGGGCCAGUUCC-CCCGACUAGAAGUCAAAUAAGUUGACAACCCAGUC---------------------GAGACCCCAAGACUCAAAGACUCUGAGACUUCAAGACCCGGCUGUC ...((((((......))-)))).......(((((.....)))))....(((((---------------------(.(.....(((.((((.......)))).))).....).)))))).. ( -27.70) >DroSim_CAF1 29205 98 - 1 AACCGGGGGCCAGUUCC-CCCGACUAGAAGUCAAAUAAGUUGACAACCCAGUC---------------------GAGACCCCAAGACUCAGAGACUCUGAGACUUCAAGACCCGGCUGUC ...((((((......))-)))).......(((((.....)))))....(((((---------------------(.(.....(((.(((((.....))))).))).....).)))))).. ( -31.70) >DroEre_CAF1 9291 106 - 1 AACCGGGGGCCAGUUCCCCCCGACUGGAACUCAAAUAAGUUGACAACCCAGCCGGGC--------------CCCGAGACCCCAGGACUCAGAGACUCUGAGACUUCAAGACCCGGCUGUC ...((((((.......))))))...((...((((.....))))...))((((((((.--------------.((.........)).(((((.....))))).........)))))))).. ( -37.70) >DroYak_CAF1 21385 120 - 1 AACCGGGGGCCACUUUCCCCUCGCCAGGAGUCAAAUAAGUUGACAACCCAGCCGAGUCCCGAGUACCGAGUCCCGAGACCCCAGGACUAAGAUACUCUGAGACUUCAAGACCCGGCUGUC ....(((((......)))))..(((.((.(((......((((......)))).((((((.((((((..(((((.(......).)))))..).))))).).)))))...)))))))).... ( -39.70) >consensus AACCGGGGGCCAGUUCC_CCCGACUAGAAGUCAAAUAAGUUGACAACCCAGUC_____________________GAGACCCCAAGACUCAGAGACUCUGAGACUUCAAGACCCGGCUGUC ..((((((((.((((......))))....(((((.....)))))......))).............................(((.(((((.....))))).))).....)))))..... (-20.18 = -20.74 + 0.56)



| Location | 6,760,628 – 6,760,726 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.53 |
| Mean single sequence MFE | -46.40 |
| Consensus MFE | -35.94 |
| Energy contribution | -35.98 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6760628 98 + 27905053 GGUCUC---------------------GACUGGGUUGUCAACUUAUUUGACUCCUAGUCGGG-GGAACUGGCCCCCGGUUAUGGCAUCCACUGGUUGCCCGGGGGUCCGCAGUUGAGCUC ..((((---------------------(((((((..(((((.....))))).))))))))))-).(((((((((((((.....(((.((...)).)))))))))))...)))))...... ( -45.80) >DroSec_CAF1 24871 98 + 1 GGUCUC---------------------GACUGGGUUGUCAACUUAUUUGACUUCUAGUCGGG-GGAACUGGCCCCCGGUUAUGGCCUCCACUGGUUGCCCGGGGGUCCGCAGUUGAGCUC ..((((---------------------(((((((..(((((.....))))).))))))))))-).(((((((((((((...(((((......))))).))))))))...)))))...... ( -44.30) >DroSim_CAF1 29245 98 + 1 GGUCUC---------------------GACUGGGUUGUCAACUUAUUUGACUUCUAGUCGGG-GGAACUGGCCCCCGGUUAUGGCCUCCACUGGUUGCCCGGGGGUCCGCAGUUGAGCUC ..((((---------------------(((((((..(((((.....))))).))))))))))-).(((((((((((((...(((((......))))).))))))))...)))))...... ( -44.30) >DroEre_CAF1 9331 106 + 1 GGUCUCGGG--------------GCCCGGCUGGGUUGUCAACUUAUUUGAGUUCCAGUCGGGGGGAACUGGCCCCCGGUUAUGGCCUCCACUGGUUGCCCGAGGGUCCGCAGUUGAGCUC ((.((((((--------------(((((((((((...((((.....))))..))))))))(((((......)))))......)))).))((((...(((....)))...)))).))))). ( -45.40) >DroYak_CAF1 21425 120 + 1 GGUCUCGGGACUCGGUACUCGGGACUCGGCUGGGUUGUCAACUUAUUUGACUCCUGGCGAGGGGAAAGUGGCCCCCGGUUAUGGCCUCCACUGGUUGCCCGGGGGUCCGCAGUUGAGCUC (((((((((........)))))))))..((..((..(((((.....))))).))..))(((..((..(((((((((((...(((((......))))).))))))).))))..))...))) ( -52.20) >consensus GGUCUC_____________________GACUGGGUUGUCAACUUAUUUGACUUCUAGUCGGG_GGAACUGGCCCCCGGUUAUGGCCUCCACUGGUUGCCCGGGGGUCCGCAGUUGAGCUC ((.(((.....................(((((((..(((((.....))))).)))))))......(((((((((((((...(((((......))))).))))))))...)))))))))). (-35.94 = -35.98 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:16 2006