
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,759,671 – 6,759,775 |
| Length | 104 |
| Max. P | 0.598719 |

| Location | 6,759,671 – 6,759,775 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -16.58 |
| Energy contribution | -15.98 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6759671 104 + 27905053 GUGG--CCCACCACCCUUCGACAUUCGUAUGCAUUCUACCCCCUAUUUGCAAGGGUGGCCAUAUUAAUGUGCCAACAGCUCUCUCGGCUUCGUGAACACACAC--------------UAC ((((--((....(((((.((.....))..((((..............))))))))))))))).....((((...((((((.....))))..))...))))...--------------... ( -23.94) >DroSec_CAF1 23893 99 + 1 GUGG--CCCACC---CCUCGACCAUCGUAUGCAUUCCACCUCCUAUUUGCGGGGGUGGCCAUAUUAAUGUGCCAACAGCUCUCUCGGCUUCGUGAACA--CAC--------------UAC ((((--((.(((---(((((.....))...(((..............))))))))))))))).....((((...((((((.....))))..))...))--)).--------------... ( -28.64) >DroSim_CAF1 28268 92 + 1 GUGG--CCCACC---CCU-------CGUAUGCAUUCCACCCCCUAUUUGCGGGGGUGGCCAUAUUAAUGUGCCAACAGCUCUCUCGGCUUCGUGAACA--CAC--------------UAC ((((--((.(((---((.-------((((..................))))))))))))))).....((((...((((((.....))))..))...))--)).--------------... ( -29.57) >DroEre_CAF1 8431 99 + 1 GUU--CCCCACC---CUUCGCACCCCAUAUGCAUUUAGCCCCCUAUUUGCGGGGGUGGCCGUAUUAAUGGGCCAACAGCUCUCUCGGCCCCAUGAACA--CAC--------------UAC (((--(.(((((---((.((((........((.....))........)))))))))))..........(((((............)))))...)))).--...--------------... ( -30.49) >DroYak_CAF1 20378 115 + 1 GUUGCCCCCACC---CUUCGCCCCACAUAUGCAUGUCGCCCACUAUUUGCGGGGGUGGCCAUAUUAAUGUGCCAACAGCUCUCUCGGCUCCAUGAACA--CACUACACACUACACACUAC ((..(((((...---.........((((....)))).((.........)))))))..))........((((.((..((((.....))))...))..))--)).................. ( -22.50) >consensus GUGG__CCCACC___CUUCGACCCUCGUAUGCAUUCCACCCCCUAUUUGCGGGGGUGGCCAUAUUAAUGUGCCAACAGCUCUCUCGGCUUCGUGAACA__CAC______________UAC ..........................(((((....((((((((.......)))))))).)))))..(((.(((............)))..)))........................... (-16.58 = -15.98 + -0.60)

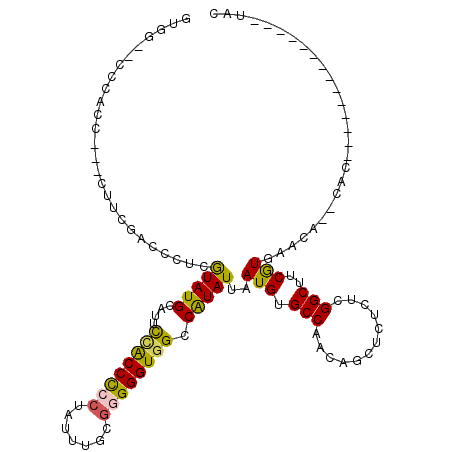

| Location | 6,759,671 – 6,759,775 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -19.54 |
| Energy contribution | -19.46 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6759671 104 - 27905053 GUA--------------GUGUGUGUUCACGAAGCCGAGAGAGCUGUUGGCACAUUAAUAUGGCCACCCUUGCAAAUAGGGGGUAGAAUGCAUACGAAUGUCGAAGGGUGGUGGG--CCAC ...--------------((((((((((((..((((....).)))..((((.(((....)))))))((((((....)))))))).))))))))))............((((....--)))) ( -34.50) >DroSec_CAF1 23893 99 - 1 GUA--------------GUG--UGUUCACGAAGCCGAGAGAGCUGUUGGCACAUUAAUAUGGCCACCCCCGCAAAUAGGAGGUGGAAUGCAUACGAUGGUCGAGG---GGUGGG--CCAC .((--------------(((--(((..((..((((....).)))))..))))))))...(((((((((((((.....(...(((.....))).)....).)).))---))).))--))). ( -35.90) >DroSim_CAF1 28268 92 - 1 GUA--------------GUG--UGUUCACGAAGCCGAGAGAGCUGUUGGCACAUUAAUAUGGCCACCCCCGCAAAUAGGGGGUGGAAUGCAUACG-------AGG---GGUGGG--CCAC ...--------------(((--.((((((...(((((.(....).))))).......((((.((((((((.......))))))))....))))..-------...---.)))))--)))) ( -36.90) >DroEre_CAF1 8431 99 - 1 GUA--------------GUG--UGUUCAUGGGGCCGAGAGAGCUGUUGGCCCAUUAAUACGGCCACCCCCGCAAAUAGGGGGCUAAAUGCAUAUGGGGUGCGAAG---GGUGGGG--AAC ...--------------.((--((((.(((((.((((.(....).))))))))).)))))).((((((.((((..(.....((.....)).....)..))))..)---)))))..--... ( -36.90) >DroYak_CAF1 20378 115 - 1 GUAGUGUGUAGUGUGUAGUG--UGUUCAUGGAGCCGAGAGAGCUGUUGGCACAUUAAUAUGGCCACCCCCGCAAAUAGUGGGCGACAUGCAUAUGUGGGGCGAAG---GGUGGGGGCAAC ....(((.(.((((.(((((--((((...(.((((....).))).).)))))))))))))..((((((((((.....))))((..(((......)))..))...)---)))))).))).. ( -37.80) >consensus GUA______________GUG__UGUUCACGAAGCCGAGAGAGCUGUUGGCACAUUAAUAUGGCCACCCCCGCAAAUAGGGGGUGGAAUGCAUACGAGGGGCGAAG___GGUGGG__CCAC ........................(((((..((((....).)))..((((.(((....)))))))(((((.......)))))...........................)))))...... (-19.54 = -19.46 + -0.08)

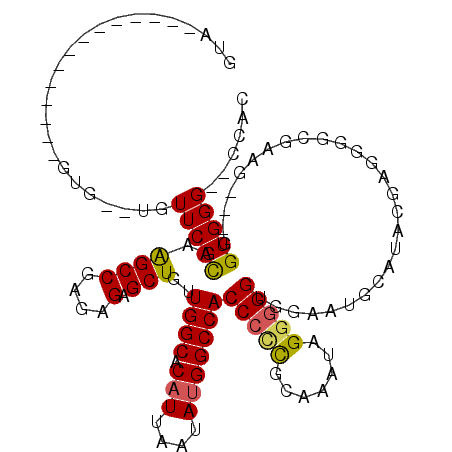
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:12 2006