
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,756,253 – 6,756,402 |
| Length | 149 |
| Max. P | 0.991379 |

| Location | 6,756,253 – 6,756,364 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -34.75 |
| Energy contribution | -34.12 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.69 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6756253 111 + 27905053 ACAAUUUCUGCCGAUGUCGCCUCAGAUGUUUAUGUUGUUGUUGCUGCUUCUGUAAAGAAGUGGGCGGCACUUUUAUGUGCCAAGGCAAAAGUUGUCGCAGCUUUAGUGGUG (((((((.((((((((((......)))))).............(..(((((....)))))..)..(((((......)))))..)))).)))))))(((.((....)).))) ( -34.90) >DroSec_CAF1 20426 111 + 1 ACAAUUUCUGCCGAUGUCGCCUCAGAUGUUUAUGUUGUUGUUGCUGCUUCUGUAAAGAAGUGGGCGGCACUUUUAUGUGCCAAGGCAAAAGUUGUCGCAGCUUUAGUGAUG .((((((.((((((((((......)))))).............(..(((((....)))))..)..(((((......)))))..)))).))))))((((.......)))).. ( -34.90) >DroSim_CAF1 24806 111 + 1 ACAAUUUCUGCCGAUGUCGCCUCAGAUGUUUAUGUUGUUGUUGCUGCUUCUGUAAAGAAGUGGGCGGCACUUUUAUGUGCCAAGGCAAAAGUUGUCGCAGCUUUAGUGAUG .((((((.((((((((((......)))))).............(..(((((....)))))..)..(((((......)))))..)))).))))))((((.......)))).. ( -34.90) >DroEre_CAF1 4864 103 + 1 ACAAUUUCUGCCGAUGUCGCCUCAGAUGUUUGU--------UGCUGCUUCUGUAAGGAAGUGGGCGGCACUUUUAUGUGCCAAGGCGAAAGUUGUCGCAGCUUUAGUGGUG .........(((((((((......)))))).((--------(((.((((((....)))))).((((((((......)))))..(((....)))))))))))......))). ( -34.00) >consensus ACAAUUUCUGCCGAUGUCGCCUCAGAUGUUUAUGUUGUUGUUGCUGCUUCUGUAAAGAAGUGGGCGGCACUUUUAUGUGCCAAGGCAAAAGUUGUCGCAGCUUUAGUGAUG .((((((.((((((((((......)))))).............(..(((((....)))))..)..(((((......)))))..)))).))))))((((.......)))).. (-34.75 = -34.12 + -0.62)



| Location | 6,756,253 – 6,756,364 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -26.94 |
| Energy contribution | -26.75 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6756253 111 - 27905053 CACCACUAAAGCUGCGACAACUUUUGCCUUGGCACAUAAAAGUGCCGCCCACUUCUUUACAGAAGCAGCAACAACAACAUAAACAUCUGAGGCGACAUCGGCAGAAAUUGU ................((((.(((((((..(((((......)))))(((..(((((....)))))(((..................))).)))......))))))).)))) ( -27.57) >DroSec_CAF1 20426 111 - 1 CAUCACUAAAGCUGCGACAACUUUUGCCUUGGCACAUAAAAGUGCCGCCCACUUCUUUACAGAAGCAGCAACAACAACAUAAACAUCUGAGGCGACAUCGGCAGAAAUUGU ................((((.(((((((..(((((......)))))(((..(((((....)))))(((..................))).)))......))))))).)))) ( -27.57) >DroSim_CAF1 24806 111 - 1 CAUCACUAAAGCUGCGACAACUUUUGCCUUGGCACAUAAAAGUGCCGCCCACUUCUUUACAGAAGCAGCAACAACAACAUAAACAUCUGAGGCGACAUCGGCAGAAAUUGU ................((((.(((((((..(((((......)))))(((..(((((....)))))(((..................))).)))......))))))).)))) ( -27.57) >DroEre_CAF1 4864 103 - 1 CACCACUAAAGCUGCGACAACUUUCGCCUUGGCACAUAAAAGUGCCGCCCACUUCCUUACAGAAGCAGCA--------ACAAACAUCUGAGGCGACAUCGGCAGAAAUUGU ...........((((((......((((((((((((......)))))((...((((......))))..)).--------..........)))))))..)).))))....... ( -27.80) >consensus CACCACUAAAGCUGCGACAACUUUUGCCUUGGCACAUAAAAGUGCCGCCCACUUCUUUACAGAAGCAGCAACAACAACAUAAACAUCUGAGGCGACAUCGGCAGAAAUUGU ...........((((((......((((((((((((......)))))((...((((......))))..))...................)))))))..)).))))....... (-26.94 = -26.75 + -0.19)



| Location | 6,756,293 – 6,756,402 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.78 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6756293 109 + 27905053 UUGCUGCUUCUGUAAAGAAGUGGGCGGCACUUUUAUGUGCCAAGGCAAAAGUUGUCGCAGCUUUAGUGGUGCCACCCAACACCACCCACCACCA--ACCCGACUUGCACCG .(((.((((((....))))))(((.(((((......)))))..((..(((((((...))))))).((((((........))))))...))....--.))).....)))... ( -36.50) >DroSec_CAF1 20466 111 + 1 UUGCUGCUUCUGUAAAGAAGUGGGCGGCACUUUUAUGUGCCAAGGCAAAAGUUGUCGCAGCUUUAGUGAUGCGACCCAACACCACCCACCACAACCACCCGACUUGCACCG .(((.((((((....))))))(((.(((((......)))))..((.....(((((((((.(......).))))))..))).....))..........))).....)))... ( -33.20) >DroSim_CAF1 24846 111 + 1 UUGCUGCUUCUGUAAAGAAGUGGGCGGCACUUUUAUGUGCCAAGGCAAAAGUUGUCGCAGCUUUAGUGAUGCCACCAAACACCACCCACCAUUACCACCCGACUUGCACCG .(((.((((((....))))))(((.(((((......)))))..(((((((((((...))))))).....))))...........)))..................)))... ( -31.80) >DroEre_CAF1 4897 97 + 1 -UGCUGCUUCUGUAAGGAAGUGGGCGGCACUUUUAUGUGCCAAGGCGAAAGUUGUCGCAGCUUUAGUGGUGCCACCCAGC----------A---CCACCCCAUUUGCACCG -(((.((((((....))))))(((.(((((......)))))...((((......)))).......(((((((......))----------)---)))))))....)))... ( -38.00) >consensus UUGCUGCUUCUGUAAAGAAGUGGGCGGCACUUUUAUGUGCCAAGGCAAAAGUUGUCGCAGCUUUAGUGAUGCCACCCAACACCACCCACCAC_ACCACCCGACUUGCACCG .(((.((((((....))))))(((.(((((......)))))..(((((((((((...))))))).....))))........................))).....)))... (-29.90 = -29.78 + -0.12)

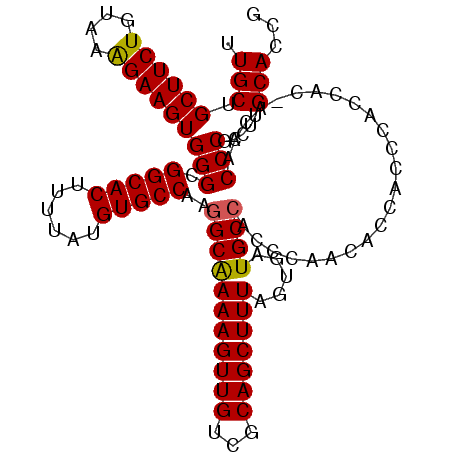

| Location | 6,756,293 – 6,756,402 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -29.73 |
| Energy contribution | -30.97 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6756293 109 - 27905053 CGGUGCAAGUCGGGU--UGGUGGUGGGUGGUGUUGGGUGGCACCACUAAAGCUGCGACAACUUUUGCCUUGGCACAUAAAAGUGCCGCCCACUUCUUUACAGAAGCAGCAA .((((.((((...((--(.(..((.(((((((((....)))))))))...))..)))).)))).))))..(((((......)))))((...(((((....)))))..)).. ( -40.30) >DroSec_CAF1 20466 111 - 1 CGGUGCAAGUCGGGUGGUUGUGGUGGGUGGUGUUGGGUCGCAUCACUAAAGCUGCGACAACUUUUGCCUUGGCACAUAAAAGUGCCGCCCACUUCUUUACAGAAGCAGCAA ...(((.....((((.((((..((.((((((((......))))))))...))..))))............(((((......))))))))).(((((....)))))..))). ( -42.20) >DroSim_CAF1 24846 111 - 1 CGGUGCAAGUCGGGUGGUAAUGGUGGGUGGUGUUUGGUGGCAUCACUAAAGCUGCGACAACUUUUGCCUUGGCACAUAAAAGUGCCGCCCACUUCUUUACAGAAGCAGCAA ...(((.....(((((((((.((((.(..((.(((((((....)))))))))..)..).))).)))))..(((((......))))))))).(((((....)))))..))). ( -39.50) >DroEre_CAF1 4897 97 - 1 CGGUGCAAAUGGGGUGG---U----------GCUGGGUGGCACCACUAAAGCUGCGACAACUUUCGCCUUGGCACAUAAAAGUGCCGCCCACUUCCUUACAGAAGCAGCA- .(.(((...((((((((---(----------(((....)))))))).......((((......))))...(((((......))))).)))).(((......)))))).).- ( -36.90) >consensus CGGUGCAAGUCGGGUGGU_GUGGUGGGUGGUGUUGGGUGGCACCACUAAAGCUGCGACAACUUUUGCCUUGGCACAUAAAAGUGCCGCCCACUUCUUUACAGAAGCAGCAA .(.(((.....((((..........(((((((((....)))))))))......((((......))))...(((((......)))))))))..(((......)))))).).. (-29.73 = -30.97 + 1.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:01 2006