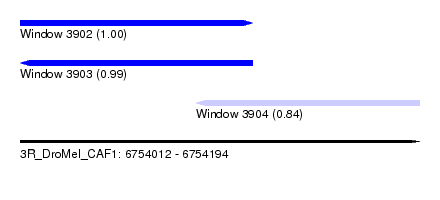
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,754,012 – 6,754,194 |
| Length | 182 |
| Max. P | 0.999973 |

| Location | 6,754,012 – 6,754,118 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.15 |
| Mean single sequence MFE | -37.54 |
| Consensus MFE | -34.66 |
| Energy contribution | -34.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.92 |
| SVM decision value | 5.09 |
| SVM RNA-class probability | 0.999973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6754012 106 + 27905053 CAAGUUAAGCAUCUCCUGGGAAGCGGAAGCAGCAGCAGAUGUUCCAGCGCCCAUUGUCCUUCAUCAGUGCCGUUGCUUCCGUUUCCUGCGACUCU--------------UUUUUUUCUUG ((((..(((....((.(((((((((((((((((.(((((((.....(((.....)))....))))..))).))))))))))))))))).))....--------------..)))..)))) ( -36.90) >DroSec_CAF1 18178 105 + 1 CAACUUAAGCAUCUCCUGGGAAGCGGAAGCAGCAGCAGAUGUUCCAGUGCCCAUUGUCCUUCAUCAGUGCCGUUGCUUCCGUUUCCUGCGACUCU--------------UUU-UUUCUUG .............((.(((((((((((((((((.(((((((...((((....)))).....))))..))).))))))))))))))))).))....--------------...-....... ( -37.10) >DroSim_CAF1 22579 105 + 1 CAACUUAAGCAUCUCCUGGGAAGCGGAAGCAGCAGCAGAUGUUCCAGUGCCCAUUGUCCUUCAUCAGUGCCGUUGCUUCCGUUUCCUGCGACUCU--------------UUU-UUUCUUG .............((.(((((((((((((((((.(((((((...((((....)))).....))))..))).))))))))))))))))).))....--------------...-....... ( -37.10) >DroEre_CAF1 2554 113 + 1 CAACUUAAGCAUCUCCUGGGAAGCGGAAGCAGCAGCAGAUGUCCCAGCGCCCAUUGUCCUUCAUCAGUGCUGUAGCUUCCGUUUCCUGCGACUCA------AUUUUUGUUUU-UUUUCUG .............((.((((((((((((((.((((((((((.....(((.....)))....))))..)))))).)))))))))))))).))....------...........-....... ( -36.40) >DroYak_CAF1 14484 119 + 1 CAACUUAAGCAUCUCCUGGGAAGCGGAAGCAGCAGCAGAUGUCCCAGCGCCCAUUGUCCUUCAUCAGUGCUGUUGCUUCCGUUUCCUGUGACUCUUCUUCUUUUUUCUUUUU-UUUUUUG .............((.(((((((((((((((((((((((((.....(((.....)))....))))..))))))))))))))))))))).)).....................-....... ( -40.20) >consensus CAACUUAAGCAUCUCCUGGGAAGCGGAAGCAGCAGCAGAUGUUCCAGCGCCCAUUGUCCUUCAUCAGUGCCGUUGCUUCCGUUUCCUGCGACUCU______________UUU_UUUCUUG .............((.(((((((((((((((((.(((((((....((.((.....)).)).))))..))).))))))))))))))))).))............................. (-34.66 = -34.86 + 0.20)



| Location | 6,754,012 – 6,754,118 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.15 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.66 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6754012 106 - 27905053 CAAGAAAAAAA--------------AGAGUCGCAGGAAACGGAAGCAACGGCACUGAUGAAGGACAAUGGGCGCUGGAACAUCUGCUGCUGCUUCCGCUUCCCAGGAGAUGCUUAACUUG ..........(--------------((.(((.(.((((.((((((((.(((((..((((..((.(....)...))....))))))))).)))))))).))))...).))).)))...... ( -31.80) >DroSec_CAF1 18178 105 - 1 CAAGAAA-AAA--------------AGAGUCGCAGGAAACGGAAGCAACGGCACUGAUGAAGGACAAUGGGCACUGGAACAUCUGCUGCUGCUUCCGCUUCCCAGGAGAUGCUUAAGUUG .......-..(--------------((.(((.(.((((.((((((((.(((((..((((.((..(.....)..))....))))))))).)))))))).))))...).))).)))...... ( -32.80) >DroSim_CAF1 22579 105 - 1 CAAGAAA-AAA--------------AGAGUCGCAGGAAACGGAAGCAACGGCACUGAUGAAGGACAAUGGGCACUGGAACAUCUGCUGCUGCUUCCGCUUCCCAGGAGAUGCUUAAGUUG .......-..(--------------((.(((.(.((((.((((((((.(((((..((((.((..(.....)..))....))))))))).)))))))).))))...).))).)))...... ( -32.80) >DroEre_CAF1 2554 113 - 1 CAGAAAA-AAAACAAAAAU------UGAGUCGCAGGAAACGGAAGCUACAGCACUGAUGAAGGACAAUGGGCGCUGGGACAUCUGCUGCUGCUUCCGCUUCCCAGGAGAUGCUUAAGUUG .......-....(((...(------(((((..(.((((.(((((((..(((((..((((..((.(....)...))....)))))))))..))))))).))))...)....)))))).))) ( -31.70) >DroYak_CAF1 14484 119 - 1 CAAAAAA-AAAAAGAAAAAAGAAGAAGAGUCACAGGAAACGGAAGCAACAGCACUGAUGAAGGACAAUGGGCGCUGGGACAUCUGCUGCUGCUUCCGCUUCCCAGGAGAUGCUUAAGUUG .......-................(((.(((.(.((((.((((((((.(((((..((((..((.(....)...))....))))))))).)))))))).))))...).))).)))...... ( -32.10) >consensus CAAGAAA_AAA______________AGAGUCGCAGGAAACGGAAGCAACGGCACUGAUGAAGGACAAUGGGCGCUGGAACAUCUGCUGCUGCUUCCGCUUCCCAGGAGAUGCUUAAGUUG ............................(((.(.((((.((((((((.(((((..((((.....((........))...))))))))).)))))))).))))...).))).......... (-29.70 = -29.66 + -0.04)



| Location | 6,754,092 – 6,754,194 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6754092 102 - 27905053 UGGCGAAACAAUAAGUGGAAAGUGCGAAAGAAAGCGAGCUGGCCAACUGGAUUGAGAUGGAGUCUAAGAUGUUGGC----CAAGAAAAAAA--------AGAGUCGCAGGAAAC ..((((..(....(((.....((.(....)...))..)))(((((((((((((.......))))))....))))))----)..........--------.)..))))....... ( -25.20) >DroSec_CAF1 18258 101 - 1 UGGCGAAACAAUAAGUGGAAAGUGCGAAAGAAAGCGAGCUGGGCAACUGGAUUGAGAUGGAGUCUAAGAUGUUGGC----CAAGAAA-AAA--------AGAGUCGCAGGAAAC ..((((..(....(((.....((.(....)...))..)))((.((((((((((.......))))))....)))).)----)......-...--------.)..))))....... ( -20.60) >DroSim_CAF1 22659 101 - 1 UGGCGAAACAAUAAGUGGAAAGUGCGAAAGAAAGCGAGCUGGCCAACUGGAUUGAGAUGGAGUCUAAGAUGUUGGC----CAAGAAA-AAA--------AGAGUCGCAGGAAAC ..((((..(....(((.....((.(....)...))..)))(((((((((((((.......))))))....))))))----)......-...--------.)..))))....... ( -25.20) >DroEre_CAF1 2634 113 - 1 UGGCGAAACAAUAAGUGGAAAGUGCGAUAGAAAGCGAGCUGGCCAACUGGAUUGAGAUGGAGUCUAAGACGUUGGUGGGACAGAAAA-AAAACAAAAAUUGAGUCGCAGGAAAC ..((((..((((...((.....(((........))).(((.((((((((((((.......))))))....)))))).)).)......-....))...))))..))))....... ( -25.20) >consensus UGGCGAAACAAUAAGUGGAAAGUGCGAAAGAAAGCGAGCUGGCCAACUGGAUUGAGAUGGAGUCUAAGAUGUUGGC____CAAGAAA_AAA________AGAGUCGCAGGAAAC ..((((..(....(((.....((.(....)...))..))).((((((((((((.......))))))....))))))........................)..))))....... (-17.59 = -17.65 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:53 2006