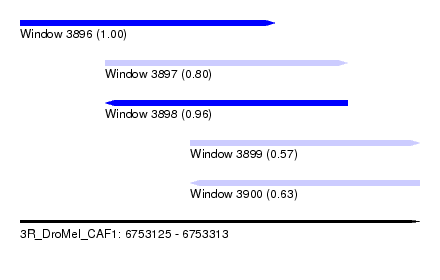
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,753,125 – 6,753,313 |
| Length | 188 |
| Max. P | 0.995152 |

| Location | 6,753,125 – 6,753,245 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -27.24 |
| Energy contribution | -27.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6753125 120 + 27905053 AAGCCCAACACCCGUCAGCAGAAUCAGCAACAACUGGCGGGCAGCUACUUACUUAAAAAGCAUUGAGCCGAAGUAAACUUUUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAG .............((((((....((((......)))).((((.(((............)))((((.((.((((....))))...)))))).......))))..))))))........... ( -27.50) >DroSec_CAF1 17293 119 + 1 AAGCCCAACACCCGUCAGCAGAAUCAGCAACAACUGGCGGGCAGCUACUUACUUAAAAAGCAUUGAGCCUAAGUAAACUUUUACGCCAAUCC-CCACGCCCACGCUGACUGCACUUAAAG .............((((((....((((......)))).((((.(((............)))((((.((.((((......)))).))))))..-....))))..))))))........... ( -27.70) >DroSim_CAF1 21693 120 + 1 AAGCCCAACACCCGUCAGCAGAAUCAGCAACAACUGGCGGGCAGCUACUUACUUAAAAAGCAUUGAGCCUAAGUAAACUUUUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAG .............((((((....((((......)))).((((.(((............)))((((.((.((((......)))).)))))).......))))..))))))........... ( -27.70) >DroEre_CAF1 1652 120 + 1 AAGCCCAACACCCGUCAGCAGAAUCAGCAACAACUGGCGGGCAGCUACUUACUUAAAAAGCAUUGAGCCUAAGUAAACUUUUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAG .............((((((....((((......)))).((((.(((............)))((((.((.((((......)))).)))))).......))))..))))))........... ( -27.70) >DroYak_CAF1 13563 120 + 1 AAGCCCAACACCCGUCAGCAGAAUCAGCAACAACUGGCGGGCAGCUACUUACUUAAAAAGCAUUGAGCCUAAGUAAACUUUUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAG .............((((((....((((......)))).((((.(((............)))((((.((.((((......)))).)))))).......))))..))))))........... ( -27.70) >consensus AAGCCCAACACCCGUCAGCAGAAUCAGCAACAACUGGCGGGCAGCUACUUACUUAAAAAGCAUUGAGCCUAAGUAAACUUUUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAG .............((((((....((((......)))).((((.(((............)))((((.((.((((......)))).)))))).......))))..))))))........... (-27.24 = -27.44 + 0.20)

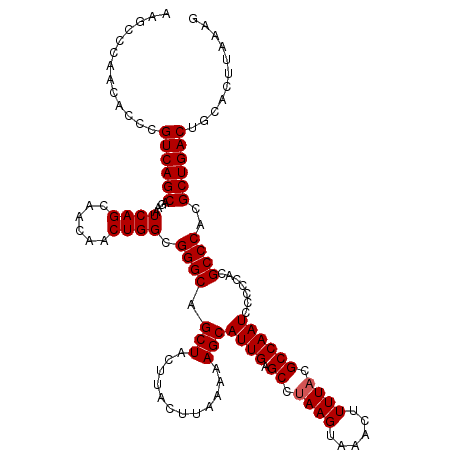

| Location | 6,753,165 – 6,753,279 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.07 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -30.12 |
| Energy contribution | -30.92 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6753165 114 + 27905053 GCAGCUACUUACUUAAAAAGCAUUGAGCCGAAGUAAACUUUUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCGUGGGGGUG------ ...(((............)))((((.((.((((....))))...))))))((((((((..(((((.....)).......(((((.......)))))..).))..))))))))..------ ( -34.40) >DroSec_CAF1 17333 113 + 1 GCAGCUACUUACUUAAAAAGCAUUGAGCCUAAGUAAACUUUUACGCCAAUCC-CCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCGUGGGGGUG------ .....(((((((((((......)))))..)))))).............((((-(((((..(((((.....)).......(((((.......)))))..).))..))))))))).------ ( -33.60) >DroSim_CAF1 21733 114 + 1 GCAGCUACUUACUUAAAAAGCAUUGAGCCUAAGUAAACUUUUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCGUGGGGGUG------ ...(((............)))((((.((.((((......)))).))))))((((((((..(((((.....)).......(((((.......)))))..).))..))))))))..------ ( -34.60) >DroEre_CAF1 1692 120 + 1 GCAGCUACUUACUUAAAAAGCAUUGAGCCUAAGUAAACUUUUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCAUGGUGGUGAAAGUG .....(((((((((((......)))))..)))))).((((((((((((...............((.....)).......(((((.......)))))..........)))).)))))))). ( -29.40) >DroYak_CAF1 13603 118 + 1 GCAGCUACUUACUUAAAAAGCAUUGAGCCUAAGUAAACUUUUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCGUGGGGGGCAGUG-- ((.(((............)))((((.((.((((......)))).))))))((((((((..(((((.....)).......(((((.......)))))..).))..))))))))))....-- ( -37.10) >consensus GCAGCUACUUACUUAAAAAGCAUUGAGCCUAAGUAAACUUUUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCGUGGGGGUG______ ...(((............)))((((.((.((((......)))).))))))((((((((..(((((.....)).......(((((.......)))))..).))..))))))))........ (-30.12 = -30.92 + 0.80)


| Location | 6,753,165 – 6,753,279 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.07 |
| Mean single sequence MFE | -40.21 |
| Consensus MFE | -38.56 |
| Energy contribution | -39.00 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6753165 114 - 27905053 ------CACCCCCACGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGGGGGAUUGGCGUAAAAGUUUACUUCGGCUCAAUGCUUUUUAAGUAAGUAGCUGC ------..((((((((..(((((((.((((((....)))))).)))).((.....)))))..))))))))...(((((..(((....)))..))....(((((.......)))))))).. ( -41.10) >DroSec_CAF1 17333 113 - 1 ------CACCCCCACGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGG-GGAUUGGCGUAAAAGUUUACUUAGGCUCAAUGCUUUUUAAGUAAGUAGCUGC ------...(((((((..(((((((.((((((....)))))).)))).((.....)))))..)))))-))...(((.......(((((((((((.....)))...))))))))..))).. ( -38.80) >DroSim_CAF1 21733 114 - 1 ------CACCCCCACGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGGGGGAUUGGCGUAAAAGUUUACUUAGGCUCAAUGCUUUUUAAGUAAGUAGCUGC ------..((((((((..(((((((.((((((....)))))).)))).((.....)))))..))))))))...(((.......(((((((((((.....)))...))))))))..))).. ( -42.30) >DroEre_CAF1 1692 120 - 1 CACUUUCACCACCAUGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGGGGGAUUGGCGUAAAAGUUUACUUAGGCUCAAUGCUUUUUAAGUAAGUAGCUGC ..((..(((..(((((.....((((.((((((....)))))).)))).........)))))..)))..))...(((.......(((((((((((.....)))...))))))))..))).. ( -35.34) >DroYak_CAF1 13603 118 - 1 --CACUGCCCCCCACGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGGGGGAUUGGCGUAAAAGUUUACUUAGGCUCAAUGCUUUUUAAGUAAGUAGCUGC --..((((((((((((..(((((((.((((((....)))))).)))).((.....)))))..))))))))..............((((((((((.....)))...))))))))))).... ( -43.50) >consensus ______CACCCCCACGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGGGGGAUUGGCGUAAAAGUUUACUUAGGCUCAAUGCUUUUUAAGUAAGUAGCUGC ........((((((((..(((((((.((((((....)))))).)))).((.....)))))..))))))))...(((.......(((((((((((.....)))...))))))))..))).. (-38.56 = -39.00 + 0.44)

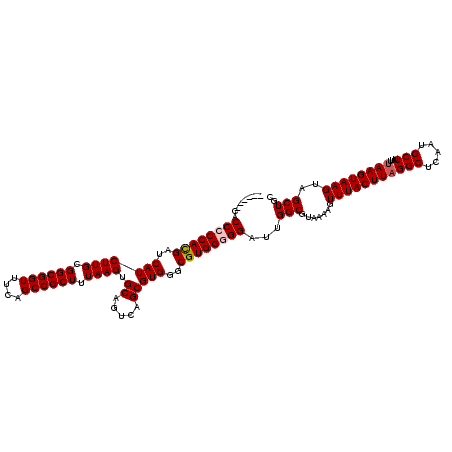
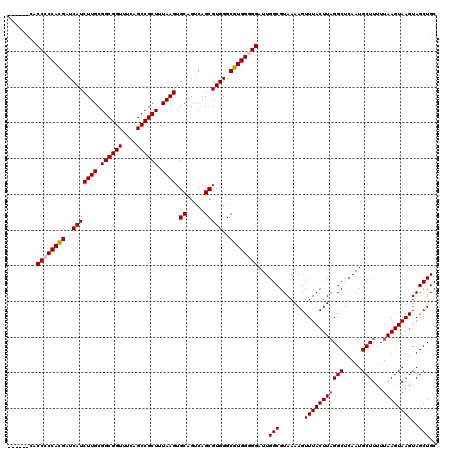
| Location | 6,753,205 – 6,753,313 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.42 |
| Mean single sequence MFE | -46.66 |
| Consensus MFE | -30.77 |
| Energy contribution | -32.57 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6753205 108 + 27905053 UUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCGUGGGGGUG------------GUGAAUGUUGGGUUGGGGUUGGGGUGCAUAUGAC .(((.((((((((((((.(((((((.....)).......(((((.......))))).........))))).)))------------).............)))))))).)))........ ( -45.01) >DroSec_CAF1 17373 107 + 1 UUACGCCAAUCC-CCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCGUGGGGGUG------------GUGAAGGUUGGGCUGGGGUUGGGGUGGAGAUGAC ((((.((((.((-(((.(((((((((............))))(((...((((((.(..(.....)...).))))------------))...))))))))))))))))).))))....... ( -47.00) >DroSim_CAF1 21773 108 + 1 UUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCGUGGGGGUG------------GUGAAGGUUGGGCUGGGGUUGGGGUGGAGAUGAC ((((.((((((((((((.(((((((.....)).......(((((.......))))).........))))).)))------------)....((.....)))))))))).))))....... ( -47.10) >DroEre_CAF1 1732 117 + 1 UUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCAUGGUGGUGAAAGUGGUGAUGCUGAAGGUUGG-C-GGGGUU-GGGUGGAGAUGAC ......((.(((((((.((((.(((..(((.........((.(((...(((((((...........)))))))...))).)).........)))..)-)-))))))-))).)))..)).. ( -46.07) >DroYak_CAF1 13643 109 + 1 UUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCGUGGGGGGCAGUG--------UGGAAGUGUGG-C-AGUGUU-GGGUGGCGAUGAC ...(((((..(((((((((((((((((.((.(((.....(((((.......)))))..(.....)))).))..)))))--------)))..))))))-.-......-))))))))..... ( -48.11) >consensus UUACGCCAAUCCCCCACGCCCACGCUGACUGCACUUAAAGCGGCUGAAACCGCCGCAAGAUGAUCGUGGGGGUG____________GUGAAGGUUGGGCUGGGGUUGGGGUGGAGAUGAC .(((.((((((((.(((.(((((((.....)).......(((((.......))))).........))))).)))..........................)))))))).)))........ (-30.77 = -32.57 + 1.80)

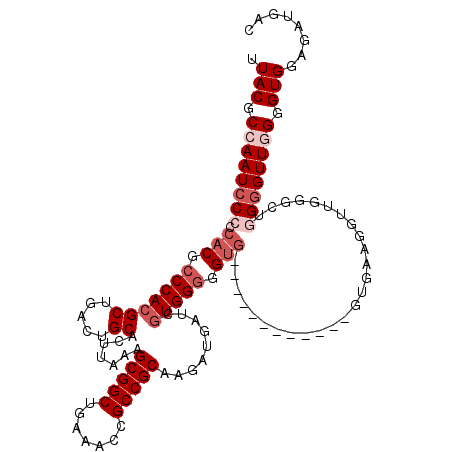
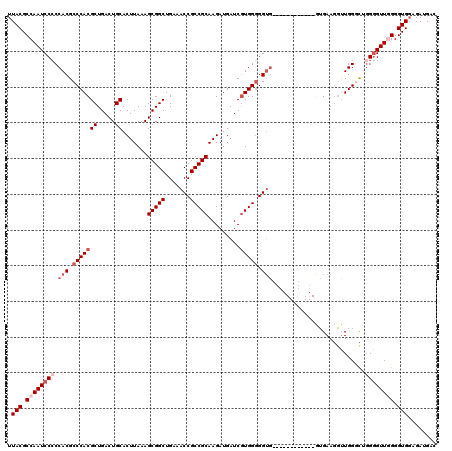
| Location | 6,753,205 – 6,753,313 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.42 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -30.62 |
| Energy contribution | -30.86 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6753205 108 - 27905053 GUCAUAUGCACCCCAACCCCAACCCAACAUUCAC------------CACCCCCACGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGGGGGAUUGGCGUAA .........((.((((.(((.............(------------(((.((((((.....((((.((((((....)))))).)))).........)))))).))))))).)))).)).. ( -37.15) >DroSec_CAF1 17373 107 - 1 GUCAUCUCCACCCCAACCCCAGCCCAACCUUCAC------------CACCCCCACGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGG-GGAUUGGCGUAA .........((.((((((((((((((........------------...............((((.((((((....)))))).)))).((.....)).))))).)))-)).)))).)).. ( -37.90) >DroSim_CAF1 21773 108 - 1 GUCAUCUCCACCCCAACCCCAGCCCAACCUUCAC------------CACCCCCACGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGGGGGAUUGGCGUAA .........((.((((.(((((......))...(------------(((.((((((.....((((.((((((....)))))).)))).........)))))).))))))).)))).)).. ( -38.54) >DroEre_CAF1 1732 117 - 1 GUCAUCUCCACCC-AACCCC-G-CCAACCUUCAGCAUCACCACUUUCACCACCAUGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGGGGGAUUGGCGUAA .........((((-((.(((-(-(.........))....((((.....((((..((((...((((.((((((....)))))).))))....))))..))))..))))))).)))).)).. ( -37.70) >DroYak_CAF1 13643 109 - 1 GUCAUCGCCACCC-AACACU-G-CCACACUUCCA--------CACUGCCCCCCACGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGGGGGAUUGGCGUAA .....(((((...-......-(-(..........--------....))((((((((..(((((((.((((((....)))))).)))).((.....)))))..))))))))..)))))... ( -39.34) >consensus GUCAUCUCCACCCCAACCCCAGCCCAACCUUCAC____________CACCCCCACGAUCAUCUUGCGGCGGUUUCAGCCGCUUUAAGUGCAGUCAGCGUGGGCGUGGGGGAUUGGCGUAA ................................................((((((((..(((((((.((((((....)))))).)))).((.....)))))..)))))))).......... (-30.62 = -30.86 + 0.24)

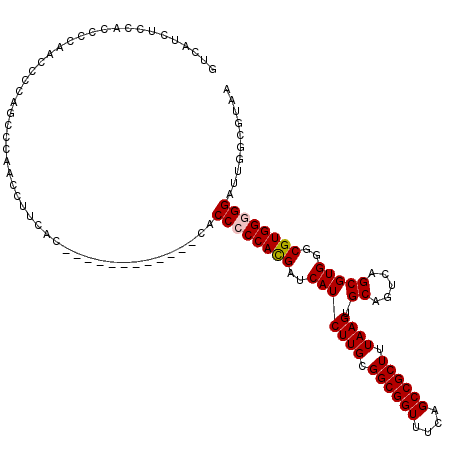
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:50 2006