
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
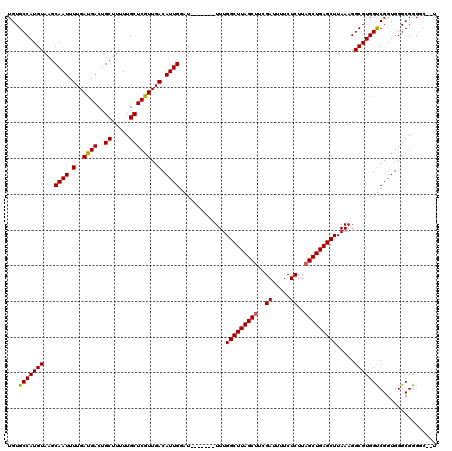
| Location | 6,752,412 – 6,752,557 |
| Length | 145 |
| Max. P | 0.985872 |

| Location | 6,752,412 – 6,752,523 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.58 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6752412 111 + 27905053 UGUGCCAUGUAAGCAAUUUUGACGACUGCUUUUUGCUCGUUGACAUUGGAU-------UUUGGCUUAGCUUCGAUUUUCUCUUAGCUGAGCUUAAAGGCGUGGUCGGUGGGCGGGGC--U ...(((....(((((.((.....)).))))).(((((((..(((..((..(-------((.(((((((((..((.....))..))))))))).)))..))..)))..))))))))))--. ( -37.00) >DroSec_CAF1 16606 111 + 1 UGUGCCAUGUAAGCAAUUUUGAUGACUGCUUUUUGCUCGUUGACAUUGGAU-------UUUGGCUUAGCUUCGAUUUUCUCUUUGCUGAGCUUAAAGGCGUGGUCGGUGGGCGAGGC--U ...(((....(((((.((.....)).))))).(((((((..(((..((..(-------((.((((((((...((.....))...)))))))).)))..))..)))..))))))))))--. ( -36.50) >DroSim_CAF1 20978 111 + 1 UGUGCCAUGUAAGCAAUUUUGAUGACUGCUUUUUGCUCGUUGACAUUGGAU-------UUUGGCUUAGCUUCGAUUUUCUCUUAGCUGAGCUUAAAGGCGUGGUCGGUGGGCGGGGC--U ...(((....(((((.((.....)).))))).(((((((..(((..((..(-------((.(((((((((..((.....))..))))))))).)))..))..)))..))))))))))--. ( -37.70) >DroEre_CAF1 896 113 + 1 UGUGCCAUGUAAGCAAUUUUGAUGACUGCUUUUUGCUCGUUGACAUUGGAU-------UUUGGCUUAGCUUCGAUUUUCUCUUAGCUGAGCUUAAAGGCGUGGCCGGUGGGCGGGCUACC ...(((.......((((.(..((((..((.....))))))..).))))..(-------((.(((((((((..((.....))..))))))))).))))))((((((.(....).)))))). ( -39.40) >DroYak_CAF1 12898 118 + 1 UGUGCCAUGUAAGCAAUUUUGAUGACUGCUUUUUGCUCGUUGACAUUGGAUUUUGGAUUUUGGCUUAGCUUCGAUUUUCUCUUAGCUGAGCUUAAAGGCGUGGUCGGUGGGCGGGCC--U ...(((.......((((.(..((((..((.....))))))..).)))).........(((.(((((((((..((.....))..))))))))).))))))..((((.(....).))))--. ( -34.80) >consensus UGUGCCAUGUAAGCAAUUUUGAUGACUGCUUUUUGCUCGUUGACAUUGGAU_______UUUGGCUUAGCUUCGAUUUUCUCUUAGCUGAGCUUAAAGGCGUGGUCGGUGGGCGGGGC__U ...(((((((...((((.(..((((..((.....))))))..).)))).............(((((((((..((.....))..))))))))).....)))))))................ (-30.70 = -30.58 + -0.12)


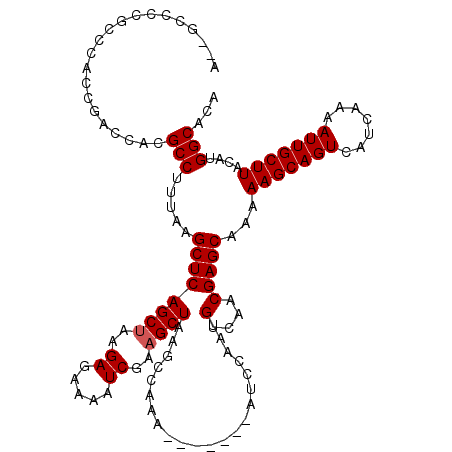
| Location | 6,752,412 – 6,752,523 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -22.89 |
| Consensus MFE | -19.70 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6752412 111 - 27905053 A--GCCCCGCCCACCGACCACGCCUUUAAGCUCAGCUAAGAGAAAAUCGAAGCUAAGCCAAA-------AUCCAAUGUCAACGAGCAAAAAGCAGUCGUCAAAAUUGCUUACAUGGCACA .--(((..((.(...(((...........(((.((((..((.....))..)))).)))....-------.......)))...).))...(((((((.......)))))))....)))... ( -22.75) >DroSec_CAF1 16606 111 - 1 A--GCCUCGCCCACCGACCACGCCUUUAAGCUCAGCAAAGAGAAAAUCGAAGCUAAGCCAAA-------AUCCAAUGUCAACGAGCAAAAAGCAGUCAUCAAAAUUGCUUACAUGGCACA .--(((..((.(...(((...........(((.(((...((.....))...))).)))....-------.......)))...).))...(((((((.......)))))))....)))... ( -21.15) >DroSim_CAF1 20978 111 - 1 A--GCCCCGCCCACCGACCACGCCUUUAAGCUCAGCUAAGAGAAAAUCGAAGCUAAGCCAAA-------AUCCAAUGUCAACGAGCAAAAAGCAGUCAUCAAAAUUGCUUACAUGGCACA .--(((..((.(...(((...........(((.((((..((.....))..)))).)))....-------.......)))...).))...(((((((.......)))))))....)))... ( -22.75) >DroEre_CAF1 896 113 - 1 GGUAGCCCGCCCACCGGCCACGCCUUUAAGCUCAGCUAAGAGAAAAUCGAAGCUAAGCCAAA-------AUCCAAUGUCAACGAGCAAAAAGCAGUCAUCAAAAUUGCUUACAUGGCACA .((.(((........))).))(((.....((((((((..((.....))..))))..(.((..-------......)).)...))))...(((((((.......)))))))....)))... ( -25.90) >DroYak_CAF1 12898 118 - 1 A--GGCCCGCCCACCGACCACGCCUUUAAGCUCAGCUAAGAGAAAAUCGAAGCUAAGCCAAAAUCCAAAAUCCAAUGUCAACGAGCAAAAAGCAGUCAUCAAAAUUGCUUACAUGGCACA .--((....))..........(((.....((((((((..((.....))..))))......................(....)))))...(((((((.......)))))))....)))... ( -21.90) >consensus A__GCCCCGCCCACCGACCACGCCUUUAAGCUCAGCUAAGAGAAAAUCGAAGCUAAGCCAAA_______AUCCAAUGUCAACGAGCAAAAAGCAGUCAUCAAAAUUGCUUACAUGGCACA .....................(((.....((((((((..((.....))..))))......................(....)))))...(((((((.......)))))))....)))... (-19.70 = -19.90 + 0.20)

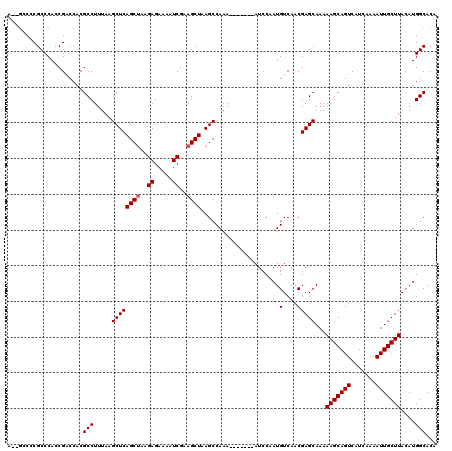
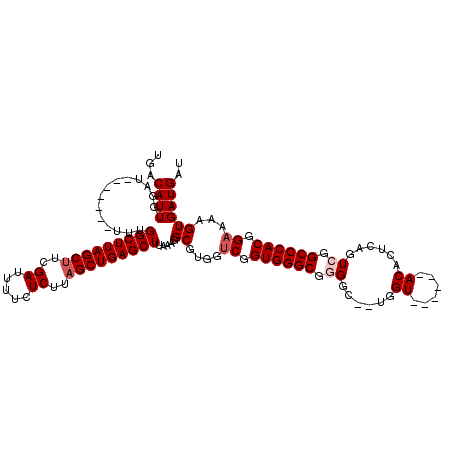
| Location | 6,752,452 – 6,752,557 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.63 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -28.56 |
| Energy contribution | -29.36 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6752452 105 + 27905053 UGACAUUGGAU-------UUUGGCUUAGCUUCGAUUUUCUCUUAGCUGAGCUUAAAGGCGUGGUCGGUGGGCGGGGC--UGGU------ACACUCAGUGGGCCCACGGAAAAGUGAUGAU ...((((...(-------((.(((((((((..((.....))..))))))))).))).((....((.((((((...((--(((.------....)))))..)))))).))...)))))).. ( -35.90) >DroSec_CAF1 16646 105 + 1 UGACAUUGGAU-------UUUGGCUUAGCUUCGAUUUUCUCUUUGCUGAGCUUAAAGGCGUGGUCGGUGGGCGAGGC--UGGU------ACACUCAGUCGGCCCACGGAAAAGUGAUGAU ...((((...(-------((.((((((((...((.....))...)))))))).))).((....((.((((((..(((--(((.------....)))))).)))))).))...)))))).. ( -37.40) >DroSim_CAF1 21018 105 + 1 UGACAUUGGAU-------UUUGGCUUAGCUUCGAUUUUCUCUUAGCUGAGCUUAAAGGCGUGGUCGGUGGGCGGGGC--UGGU------ACACUCAGUCGGCCCACGGAAAAGUGAUGAU ...((((...(-------((.(((((((((..((.....))..))))))))).))).((....((.((((((..(((--(((.------....)))))).)))))).))...)))))).. ( -38.20) >DroEre_CAF1 936 113 + 1 UGACAUUGGAU-------UUUGGCUUAGCUUCGAUUUUCUCUUAGCUGAGCUUAAAGGCGUGGCCGGUGGGCGGGCUACCAGUGUACCAACAUUCAGUCGGCCCACGGAAAAGUGAUGAU ...((((...(-------((.(((((((((..((.....))..))))))))).))).((....(((.(((((.((((...(((((....))))).)))).))))))))....)))))).. ( -39.80) >DroYak_CAF1 12938 112 + 1 UGACAUUGGAUUUUGGAUUUUGGCUUAGCUUCGAUUUUCUCUUAGCUGAGCUUAAAGGCGUGGUCGGUGGGCGGGCC--UGGU------ACAUUCGGUCGGCCCACGGAAAAGUGAUGAU ...((((.(((..((..(((.(((((((((..((.....))..))))))))).)))..))..))).((((((.((((--.((.------....)))))).))))))........)))).. ( -41.40) >consensus UGACAUUGGAU_______UUUGGCUUAGCUUCGAUUUUCUCUUAGCUGAGCUUAAAGGCGUGGUCGGUGGGCGGGGC__UGGU______ACACUCAGUCGGCCCACGGAAAAGUGAUGAU ...((((..............(((((((((..((.....))..))))))))).....((....((.((((((.((......((......))......)).)))))).))...)))))).. (-28.56 = -29.36 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:45 2006