
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,748,163 – 6,748,472 |
| Length | 309 |
| Max. P | 0.999253 |

| Location | 6,748,163 – 6,748,278 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -19.81 |
| Energy contribution | -19.62 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6748163 115 + 27905053 UUAACUUUUCAUUGCCUAGUAGGGCAAACACAAUUAGGGGCCAUCCACAAAACUAAAA-AA--AAAA--AUCGAAUAGUAGUAAAAAGACAAAAGGCAGGGGAAACUUUGACAAUUAUUU ...(((.(((.((((((....)))))).........(((....)))............-..--....--...))).)))...............(.(((((....))))).)........ ( -19.90) >DroSec_CAF1 12408 114 + 1 UUAACUUUUCAUUGCCUAGUAGGGCAAACACAAUUAGGGGCCACCCCCAAAACUAAAA-AA--AAAAACAUCGAAUA---GUAAAAAGACAAAAGGCAGGGGAAACUUUGACAAUUAUUU ...(((.(((.((((((....)))))).........((((....))))..........-..--.........))).)---))............(.(((((....))))).)........ ( -24.80) >DroSim_CAF1 16721 116 + 1 UUAACUUUUCAUUGCCUAGUAGGGCAAACACAAUUAGGGGCCACCCCCAAAACUAAAA-AAAAAAAAACAUCGAAUA---GUAAAAAGACAAAAGGCAGGGGAAACUUUGACAAUUAUUU ...(((.(((.((((((....)))))).........((((....))))..........-.............))).)---))............(.(((((....))))).)........ ( -24.80) >DroYak_CAF1 8529 117 + 1 UUAACUUUUCAUUGCCUAGUAGGGCAAACACAAUUAGGGGCCACCCGCGAAACUAAACUAAAAAAAAACAUCGAAUA---GUAAAAAGACAAAAGGCAGGGGAAACUUUGACAAUUAUUU ...(((.(((.((((((....)))))).........(((....)))..........................))).)---))............(.(((((....))))).)........ ( -23.20) >consensus UUAACUUUUCAUUGCCUAGUAGGGCAAACACAAUUAGGGGCCACCCCCAAAACUAAAA_AA__AAAAACAUCGAAUA___GUAAAAAGACAAAAGGCAGGGGAAACUUUGACAAUUAUUU ....(((((..((((((....)))))).........(((....)))............................................))))).(((((....))))).......... (-19.81 = -19.62 + -0.19)


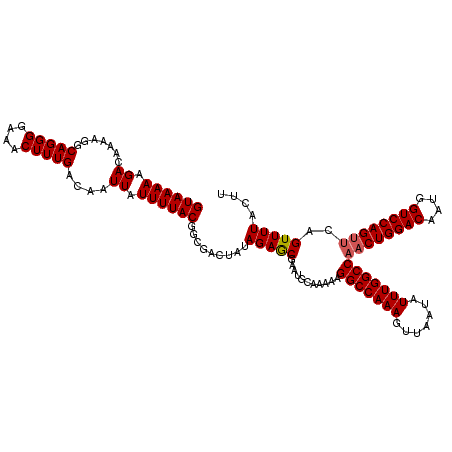
| Location | 6,748,238 – 6,748,358 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -32.14 |
| Energy contribution | -32.45 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6748238 120 + 27905053 GUAAAAAGACAAAAGGCAGGGGAAACUUUGACAAUUAUUUUACGGCGACUAUAGAGCGAAUGCAAAAAGGCCAAAGUUAAUAUUUGGCCAACUGGACAAUGGUCCAGAACAGUUUUCCUU ((((((.((.......(((((....)))))....)).))))))..........(((.(((.((.....(((((((.......)))))))..((((((....))))))....)).)))))) ( -33.50) >DroSec_CAF1 12482 120 + 1 GUAAAAAGACAAAAGGCAGGGGAAACUUUGACAAUUAUUUUACGGCGACUAUAGAACGAAUGCAAAAAGGCCAAAGUUAAUAUUUGGCCAACUGGACAAUGGUCCAGUUCAGUUUUACUU ((((((.((.......(((((....)))))....)).))))))........((((((...........(((((((.......)))))))((((((((....))))))))..))))))... ( -34.70) >DroSim_CAF1 16797 120 + 1 GUAAAAAGACAAAAGGCAGGGGAAACUUUGACAAUUAUUUUACGGCGACUAUAGAGCGAAUGCAAAAAGGCCAAAGUUAAUAUUUGGCCAACUGGACAAUGGUCCAGUUCAGUUUUACUU ((((((.((.......(((((....)))))....)).))))))........((((((...........(((((((.......)))))))((((((((....))))))))..))))))... ( -34.70) >DroYak_CAF1 8606 116 + 1 GUAAAAAGACAAAAGGCAGGGGAAACUUUGACAAUUAUUUUACGGCGACUAUAGAGCGAAUGCAAAAAGGCCAAAGUUAAUAUUUGGCCAACUGGACAAUGGUCCAGUUGAGUUUU---- ((((((.((.......(((((....)))))....)).)))))).........(((((...........(((((((.......)))))))((((((((....))))))))..)))))---- ( -33.40) >consensus GUAAAAAGACAAAAGGCAGGGGAAACUUUGACAAUUAUUUUACGGCGACUAUAGAGCGAAUGCAAAAAGGCCAAAGUUAAUAUUUGGCCAACUGGACAAUGGUCCAGUUCAGUUUUACUU ((((((.((.......(((((....)))))....)).)))))).........(((((...........(((((((.......)))))))((((((((....))))))))..))))).... (-32.14 = -32.45 + 0.31)


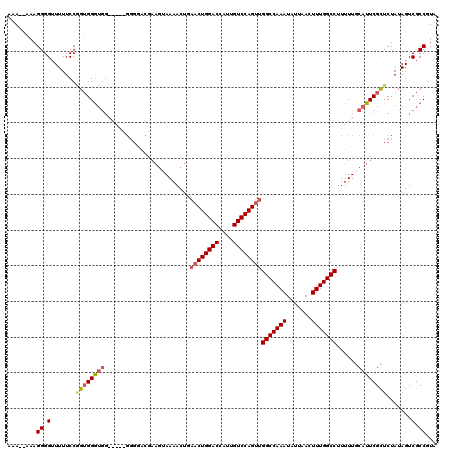
| Location | 6,748,238 – 6,748,358 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -27.19 |
| Consensus MFE | -24.85 |
| Energy contribution | -25.16 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6748238 120 - 27905053 AAGGAAAACUGUUCUGGACCAUUGUCCAGUUGGCCAAAUAUUAACUUUGGCCUUUUUGCAUUCGCUCUAUAGUCGCCGUAAAAUAAUUGUCAAAGUUUCCCCUGCCUUUUGUCUUUUUAC (((((........((((((....))))))..(((........(((((((((..((((((...((((....)).))..)))))).....)))))))))......))).....))))).... ( -25.34) >DroSec_CAF1 12482 120 - 1 AAGUAAAACUGAACUGGACCAUUGUCCAGUUGGCCAAAUAUUAACUUUGGCCUUUUUGCAUUCGUUCUAUAGUCGCCGUAAAAUAAUUGUCAAAGUUUCCCCUGCCUUUUGUCUUUUUAC ..((((((...((((((((....))))))))(((........(((((((((..((((((...((.........))..)))))).....)))))))))......)))........)))))) ( -27.84) >DroSim_CAF1 16797 120 - 1 AAGUAAAACUGAACUGGACCAUUGUCCAGUUGGCCAAAUAUUAACUUUGGCCUUUUUGCAUUCGCUCUAUAGUCGCCGUAAAAUAAUUGUCAAAGUUUCCCCUGCCUUUUGUCUUUUUAC ..((((((...((((((((....))))))))(((........(((((((((..((((((...((((....)).))..)))))).....)))))))))......)))........)))))) ( -28.84) >DroYak_CAF1 8606 116 - 1 ----AAAACUCAACUGGACCAUUGUCCAGUUGGCCAAAUAUUAACUUUGGCCUUUUUGCAUUCGCUCUAUAGUCGCCGUAAAAUAAUUGUCAAAGUUUCCCCUGCCUUUUGUCUUUUUAC ----.......((((((((....))))))))(((........(((((((((..((((((...((((....)).))..)))))).....)))))))))......))).............. ( -26.74) >consensus AAGUAAAACUGAACUGGACCAUUGUCCAGUUGGCCAAAUAUUAACUUUGGCCUUUUUGCAUUCGCUCUAUAGUCGCCGUAAAAUAAUUGUCAAAGUUUCCCCUGCCUUUUGUCUUUUUAC ...........((((((((....))))))))(((........(((((((((..((((((...((((....)).))..)))))).....)))))))))......))).............. (-24.85 = -25.16 + 0.31)



| Location | 6,748,278 – 6,748,392 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -24.12 |
| Energy contribution | -24.25 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6748278 114 + 27905053 UACGGCGACUAUAGAGCGAAUGCAAAAAGGCCAAAGUUAAUAUUUGGCCAACUGGACAAUGGUCCAGAACAGUUUUCCUUUCUUCCC------UCUCCACAGGAAAAACCCCUUUUUUUU ...((.((....((((.(((.((.....(((((((.......)))))))..((((((....))))))....)).)))..))))....------)).))..(((.......)))....... ( -29.60) >DroSec_CAF1 12522 118 + 1 UACGGCGACUAUAGAACGAAUGCAAAAAGGCCAAAGUUAAUAUUUGGCCAACUGGACAAUGGUCCAGUUCAGUUUUACUUCGUCCCCAUCCCCCACCCACCGGAAAAACCCCUUU--UUU ...(((((...((((((...........(((((((.......)))))))((((((((....))))))))..))))))..)))))....(((..........)))...........--... ( -32.40) >DroSim_CAF1 16837 118 + 1 UACGGCGACUAUAGAGCGAAUGCAAAAAGGCCAAAGUUAAUAUUUGGCCAACUGGACAAUGGUCCAGUUCAGUUUUACUUCGUCCCCAUCCCCCACCCACCGGAAAAACCCCUUU--UUU ...(((((...((((((...........(((((((.......)))))))((((((((....))))))))..))))))..)))))....(((..........)))...........--... ( -32.40) >DroYak_CAF1 8646 104 + 1 UACGGCGACUAUAGAGCGAAUGCAAAAAGGCCAAAGUUAAUAUUUGGCCAACUGGACAAUGGUCCAGUUGAGUUUU----------------CCACCCCCUGGAAAACCCCCUAGUUUUG ...........((((((...........(((((((.......)))))))((((((((....))))))))..(((((----------------(((.....))))))))......)))))) ( -33.50) >consensus UACGGCGACUAUAGAGCGAAUGCAAAAAGGCCAAAGUUAAUAUUUGGCCAACUGGACAAUGGUCCAGUUCAGUUUUACUUCGUCCCC_____CCACCCACCGGAAAAACCCCUUU__UUU ...((.(.....(((((...........(((((((.......)))))))((((((((....))))))))..)))))...................((....)).....).))........ (-24.12 = -24.25 + 0.12)



| Location | 6,748,278 – 6,748,392 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -26.61 |
| Energy contribution | -27.61 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6748278 114 - 27905053 AAAAAAAAGGGGUUUUUCCUGUGGAGA------GGGAAGAAAGGAAAACUGUUCUGGACCAUUGUCCAGUUGGCCAAAUAUUAACUUUGGCCUUUUUGCAUUCGCUCUAUAGUCGCCGUA ..........(((.....((((((((.------..(((...(((((.......((((((....))))))..(((((((.......))))))))))))...))).))))))))..)))... ( -32.30) >DroSec_CAF1 12522 118 - 1 AAA--AAAGGGGUUUUUCCGGUGGGUGGGGGAUGGGGACGAAGUAAAACUGAACUGGACCAUUGUCCAGUUGGCCAAAUAUUAACUUUGGCCUUUUUGCAUUCGUUCUAUAGUCGCCGUA ...--....(((....)))........((.(((.((((((((((((((...((((((((....))))))))(((((((.......))))))).)))))).))))))))...))).))... ( -44.00) >DroSim_CAF1 16837 118 - 1 AAA--AAAGGGGUUUUUCCGGUGGGUGGGGGAUGGGGACGAAGUAAAACUGAACUGGACCAUUGUCCAGUUGGCCAAAUAUUAACUUUGGCCUUUUUGCAUUCGCUCUAUAGUCGCCGUA ...--....(((....)))....(((((...((((((.((((((((((...((((((((....))))))))(((((((.......))))))).)))))).))))))))))..)))))... ( -43.50) >DroYak_CAF1 8646 104 - 1 CAAAACUAGGGGGUUUUCCAGGGGGUGG----------------AAAACUCAACUGGACCAUUGUCCAGUUGGCCAAAUAUUAACUUUGGCCUUUUUGCAUUCGCUCUAUAGUCGCCGUA ........((.(..(....((.(((((.----------------((((...((((((((....))))))))(((((((.......))))))).)))).))))).))....)..).))... ( -35.00) >consensus AAA__AAAGGGGUUUUUCCGGUGGGUGG_____GGGGACGAAGUAAAACUGAACUGGACCAUUGUCCAGUUGGCCAAAUAUUAACUUUGGCCUUUUUGCAUUCGCUCUAUAGUCGCCGUA ........((.(.......((((((((........................((((((((....))))))))(((((((.......)))))))......)))))))).......).))... (-26.61 = -27.61 + 1.00)


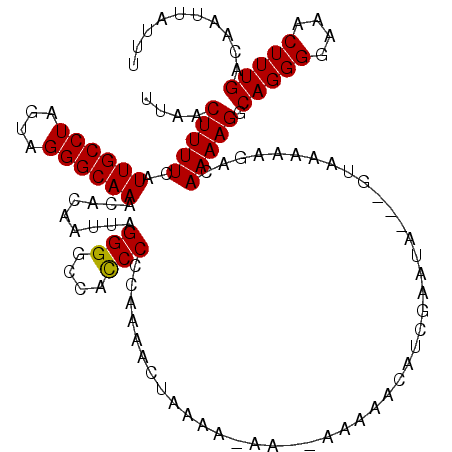
| Location | 6,748,318 – 6,748,432 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.70 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6748318 114 + 27905053 UAUUUGGCCAACUGGACAAUGGUCCAGAACAGUUUUCCUUUCUUCCC------UCUCCACAGGAAAAACCCCUUUUUUUUCGGGGACACCAGCCCACCGGCUGACUUUCCUUCAGUUUCG ...((((....((((((....))))))....................------.......((((((..((((.........))))....(((((....)))))..))))))))))..... ( -29.00) >DroSec_CAF1 12562 115 + 1 UAUUUGGCCAACUGGACAAUGGUCCAGUUCAGUUUUACUUCGUCCCCAUCCCCCACCCACCGGAAAAACCCCUUU--UUUCGGGGACACCAGCCCACCGGCUGACUUU---UCAGUUUCG .....(((.((((((((....))))))))............((((((......(.......)((((((.....))--))))))))))....)))....((((((....---))))))... ( -32.30) >DroSim_CAF1 16877 115 + 1 UAUUUGGCCAACUGGACAAUGGUCCAGUUCAGUUUUACUUCGUCCCCAUCCCCCACCCACCGGAAAAACCCCUUU--UUUCGGGGACACCAGCCCACCGGCUGACUUU---CCAGUUUCG ...((((..((((((((....))))))))............((((((......(.......)((((((.....))--))))))))))..(((((....))))).....---))))..... ( -33.70) >DroYak_CAF1 8686 101 + 1 UAUUUGGCCAACUGGACAAUGGUCCAGUUGAGUUUU----------------CCACCCCCUGGAAAACCCCCUAGUUUUGGGGGGUCACCAGCCCAACGGCUGACUUU---CCAGUUUGG .....((((((((((((....))))))))).)))..----------------(((....(((((((((((((((....))))))))...(((((....)))))..)))---))))..))) ( -44.90) >consensus UAUUUGGCCAACUGGACAAUGGUCCAGUUCAGUUUUACUUCGUCCCC_____CCACCCACCGGAAAAACCCCUUU__UUUCGGGGACACCAGCCCACCGGCUGACUUU___CCAGUUUCG ...((((..((((((((....)))))))).(((...................................((((.........))))....(((((....)))))))).....))))..... (-24.62 = -24.88 + 0.25)

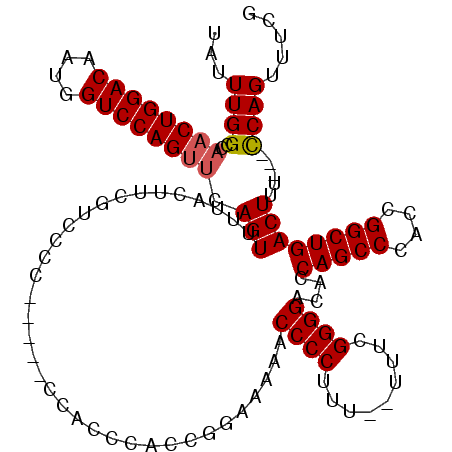
| Location | 6,748,318 – 6,748,432 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -40.03 |
| Consensus MFE | -25.75 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6748318 114 - 27905053 CGAAACUGAAGGAAAGUCAGCCGGUGGGCUGGUGUCCCCGAAAAAAAAGGGGUUUUUCCUGUGGAGA------GGGAAGAAAGGAAAACUGUUCUGGACCAUUGUCCAGUUGGCCAAAUA .....(((.(((((((((((((....))))))...((((.........)))).))))))).)))...------.((..((.((.....))...((((((....))))))))..))..... ( -32.60) >DroSec_CAF1 12562 115 - 1 CGAAACUGA---AAAGUCAGCCGGUGGGCUGGUGUCCCCGAAA--AAAGGGGUUUUUCCGGUGGGUGGGGGAUGGGGACGAAGUAAAACUGAACUGGACCAUUGUCCAGUUGGCCAAAUA ....(((..---..)))..((((((..(((..((((((((...--........((..(((.....)))..)))))))))).)))...))).((((((((....)))))))))))...... ( -38.80) >DroSim_CAF1 16877 115 - 1 CGAAACUGG---AAAGUCAGCCGGUGGGCUGGUGUCCCCGAAA--AAAGGGGUUUUUCCGGUGGGUGGGGGAUGGGGACGAAGUAAAACUGAACUGGACCAUUGUCCAGUUGGCCAAAUA ....(((((---((((((((((....))))))...((((....--...)))).))))))))).(((......((....))...........((((((((....)))))))).)))..... ( -41.90) >DroYak_CAF1 8686 101 - 1 CCAAACUGG---AAAGUCAGCCGUUGGGCUGGUGACCCCCCAAAACUAGGGGGUUUUCCAGGGGGUGG----------------AAAACUCAACUGGACCAUUGUCCAGUUGGCCAAAUA ((...((((---((..((((((....)))))).(((((((........))))))))))))).)).(((----------------.....((((((((((....))))))))))))).... ( -46.80) >consensus CGAAACUGA___AAAGUCAGCCGGUGGGCUGGUGUCCCCGAAA__AAAGGGGUUUUUCCGGUGGGUGG_____GGGGACGAAGUAAAACUGAACUGGACCAUUGUCCAGUUGGCCAAAUA ...................(((.....(((((...((((.........)))).....))))).............................((((((((....)))))))))))...... (-25.75 = -26.62 + 0.88)



| Location | 6,748,358 – 6,748,472 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -30.11 |
| Energy contribution | -30.18 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6748358 114 + 27905053 UCUUCCC------UCUCCACAGGAAAAACCCCUUUUUUUUCGGGGACACCAGCCCACCGGCUGACUUUCCUUCAGUUUCGUGGGACACAUGGCUUCGGAGCGAACUAGAGUAAUGUCGUG .......------..(((((((((((..((((.........))))....(((((....)))))..))))))........)))))((((((.((((..(......)..)))).)))).)). ( -31.30) >DroSec_CAF1 12602 115 + 1 CGUCCCCAUCCCCCACCCACCGGAAAAACCCCUUU--UUUCGGGGACACCAGCCCACCGGCUGACUUU---UCAGUUUCGUGGGACACAUGGCUUCGGAGCGAACUAGAGUAAUGUCGUG .((((((.........((.(((((((((....)))--))))))))....(((((....))))).....---........).)))))((((.((((..(......)..)))).)))).... ( -34.90) >DroSim_CAF1 16917 115 + 1 CGUCCCCAUCCCCCACCCACCGGAAAAACCCCUUU--UUUCGGGGACACCAGCCCACCGGCUGACUUU---CCAGUUUCGUGGGACACAUGGCUUCGGAGCGAACUAGAGUAAUGUCGUG ...........(((((..((.(((((..((((...--....))))....(((((....)))))..)))---)).))...)))))((((((.((((..(......)..)))).)))).)). ( -36.50) >DroYak_CAF1 8722 105 + 1 ------------CCACCCCCUGGAAAACCCCCUAGUUUUGGGGGGUCACCAGCCCAACGGCUGACUUU---CCAGUUUGGUGGGACACAUGGCUUCGGAGCGAACUAGAGUAAUGUCGUG ------------(((((..(((((((((((((((....))))))))...(((((....)))))..)))---))))...)))))((((....((((..(......)..))))..))))... ( -45.40) >consensus CGUCCCC_____CCACCCACCGGAAAAACCCCUUU__UUUCGGGGACACCAGCCCACCGGCUGACUUU___CCAGUUUCGUGGGACACAUGGCUUCGGAGCGAACUAGAGUAAUGUCGUG ...............(((((..((....((((.........))))....(((((....)))))............))..)))))((((((.((((..(......)..)))).)))).)). (-30.11 = -30.18 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:34 2006