
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,744,853 – 6,745,008 |
| Length | 155 |
| Max. P | 0.862145 |

| Location | 6,744,853 – 6,744,968 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.57 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6744853 115 - 27905053 C-----ACAAGCAUACACACACACACACCCUUUCAAUGUAGUGUGUCUGUGUGCGCAUUUCAAUAAAUCGCUCGCAUCUACAAUAACACGAAAGCAUGCGGCCAAAAGUAGGCAGCCUGC .-----....(((((((.(((((((((.........))).)))))).)))))))...............((.(((((((.............)).))))))).....(((((...))))) ( -30.32) >DroSec_CAF1 9574 111 - 1 C-----ACAAGCA----CACACGCACACCCUUUCAAUGUAGUGUGUCUGUGUGCGCAUUUCAAUAAAUCGCUCGCUUCUACAAUAACACGAAAGCAUGCGGCCAAAAGUAGGCAGCCUAC .-----....(((----((((.((((((............)))))).)))))))((............(((..(((((...........).))))..)))(((.......))).)).... ( -31.70) >DroYak_CAF1 5226 116 - 1 CCGGACCCGAGCA----CACACGCACACCCUUUCAAUGUAGUGUGUCUGUGUGCGCAUUUCAAUAAAUCGCUCGCUUCUACAAUAACACGAUAGCAUGCGGCCAAAAGUAGGCAGCCUGC ..(((..((((((----((((.((((((............)))))).)))))))((((((....)))).)))))..)))..............(((.((.(((.......))).)).))) ( -34.10) >consensus C_____ACAAGCA____CACACGCACACCCUUUCAAUGUAGUGUGUCUGUGUGCGCAUUUCAAUAAAUCGCUCGCUUCUACAAUAACACGAAAGCAUGCGGCCAAAAGUAGGCAGCCUGC ........((((.....((((.((((((............)))))).)))).(((.((((....)))))))..))))................(((.((.(((.......))).)).))) (-24.34 = -24.57 + 0.23)

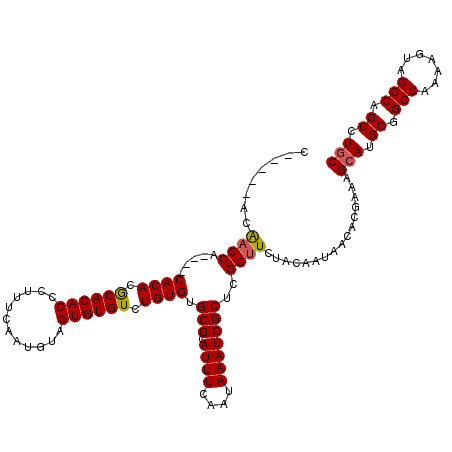

| Location | 6,744,893 – 6,745,008 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -41.93 |
| Consensus MFE | -32.11 |
| Energy contribution | -31.57 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6744893 115 + 27905053 UAGAUGCGAGCGAUUUAUUGAAAUGCGCACACAGACACACUACAUUGAAAGGGUGUGUGUGUGUGUAUGCUUGU-----GUGGUGCUUUUUGAACAACGUUUGGCUGGCUCGUCGCUUGA ......((((((((.(((....(((((((((((.((((.((........)).)))).)))))))))))....))-----)..(.(((..(..(((...)))..)..))).))))))))). ( -40.70) >DroSec_CAF1 9614 110 + 1 UAGAAGCGAGCGAUUUAUUGAAAUGCGCACACAGACACACUACAUUGAAAGGGUGUGCGUGUG----UGCUUGU-----GUGAGGCCUUUUGAACAACGUUCGGCAGGCUCGUCGCUUG- ...((((((.((((((((...((.((((((((.(.((((((..........))))))))))))----)))))..-----)))))((((.((((((...)))))).))))))))))))).- ( -41.70) >DroYak_CAF1 5266 115 + 1 UAGAAGCGAGCGAUUUAUUGAAAUGCGCACACAGACACACUACAUUGAAAGGGUGUGCGUGUG----UGCUCGGGUCCGGUGAGGCCUUUUGAACAACGUUUGGCAGGCUCGUCGCUUG- ...((((((.(((((((((((..((.(((((((..((((((..........))))))..))))----))).))..).)))))))((((.(..(((...)))..).))))))))))))).- ( -43.40) >consensus UAGAAGCGAGCGAUUUAUUGAAAUGCGCACACAGACACACUACAUUGAAAGGGUGUGCGUGUG____UGCUUGU_____GUGAGGCCUUUUGAACAACGUUUGGCAGGCUCGUCGCUUG_ ......((((((((........((((((((((.....((......)).....))))))))))...................((.((((.((((((...)))))).)))))))))))))). (-32.11 = -31.57 + -0.55)

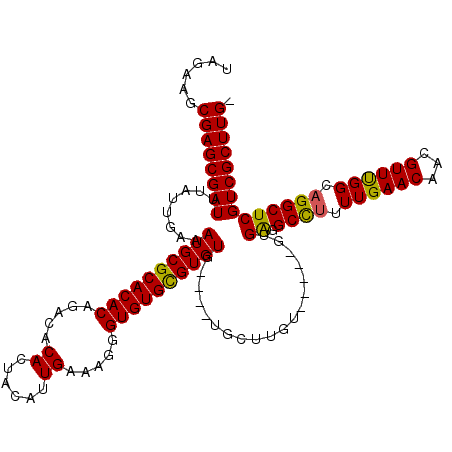

| Location | 6,744,893 – 6,745,008 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -24.85 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6744893 115 - 27905053 UCAAGCGACGAGCCAGCCAAACGUUGUUCAAAAAGCACCAC-----ACAAGCAUACACACACACACACCCUUUCAAUGUAGUGUGUCUGUGUGCGCAUUUCAAUAAAUCGCUCGCAUCUA ....(((((((..((((.....))))...............-----....(((((((.(((((((((.........))).)))))).))))))).............))).))))..... ( -30.20) >DroSec_CAF1 9614 110 - 1 -CAAGCGACGAGCCUGCCGAACGUUGUUCAAAAGGCCUCAC-----ACAAGCA----CACACGCACACCCUUUCAAUGUAGUGUGUCUGUGUGCGCAUUUCAAUAAAUCGCUCGCUUCUA -.(((((((((((((...((((...))))...)))).....-----....(((----((((.((((((............)))))).))))))).............))).))))))... ( -38.00) >DroYak_CAF1 5266 115 - 1 -CAAGCGACGAGCCUGCCAAACGUUGUUCAAAAGGCCUCACCGGACCCGAGCA----CACACGCACACCCUUUCAAUGUAGUGUGUCUGUGUGCGCAUUUCAAUAAAUCGCUCGCUUCUA -.(((((((((((((....(((...)))....)))).....((....)).(((----((((.((((((............)))))).))))))).............))).))))))... ( -35.30) >consensus _CAAGCGACGAGCCUGCCAAACGUUGUUCAAAAGGCCUCAC_____ACAAGCA____CACACGCACACCCUUUCAAUGUAGUGUGUCUGUGUGCGCAUUUCAAUAAAUCGCUCGCUUCUA ....(((((((((((....(((...)))....))))..............(((....((((.((((((............)))))).))))))).............))).))))..... (-24.85 = -25.30 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:18 2006