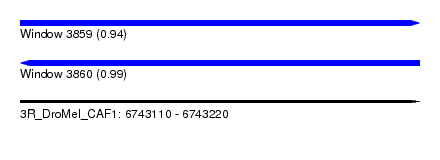
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,743,110 – 6,743,220 |
| Length | 110 |
| Max. P | 0.992190 |

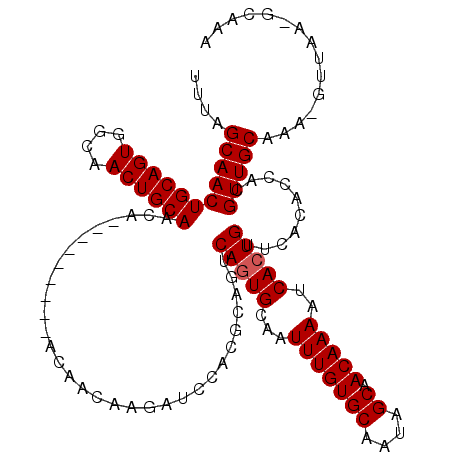
| Location | 6,743,110 – 6,743,220 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -28.47 |
| Energy contribution | -29.03 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6743110 110 + 27905053 UUUGCAUUAAC-UUUGCAACGGGGUGUGAACAGUGAUUUUGUUGCUAUUGCACAAAUUGCACUGACUGCGUGGAUCUUGUUGU---------UGUUGCAGUUGCCACUGCAGUUGCUAAA .(((((.....-..)))))...(((.....(((((..(((((.((....)))))))...)))))(((((((((...((((...---------....))))...)))).))))).)))... ( -32.50) >DroSec_CAF1 7890 109 + 1 UUCGC-UUAAC-UUUGCAACGUGGUGUGAACAGUGAUUUUGUUGCUAUUGCACAAAUUGCACUGACUGCGUGGAUCUUGUUGU---------UGUUGCAGUUGCCACUGCAGUUGCUAAA .....-.....-...((((((..(((....(((((..(((((.((....)))))))...)))))((((((..(((......))---------)..))))))...)))..).))))).... ( -33.00) >DroSim_CAF1 12229 109 + 1 UUCGC-UUAAC-UUUGCAACGUGGUGUGAACAGUGAUUUUGUUGCUAUUGCACAAAUUGCACUGACUGCGUGGAUCUUGUUGU---------UGUUGCAGUUGCCACUGCAGUUGCUAAA .....-.....-...((((((..(((....(((((..(((((.((....)))))))...)))))((((((..(((......))---------)..))))))...)))..).))))).... ( -33.00) >DroYak_CAF1 3209 117 + 1 UUUGA---AACUUUUGCAACGGAGUGUGAACAGUGAUUUUGUUGCUGUUGCACAAAUUGCAAUGACUGCGUGGAACUUGUUUUUGUUGCUGCUGUUGCAGUUGCCACUGCAGUUGCUAAA .....---.....((((((.....(((((((((..((...))..))))).))))..)))))).(((((((((((((........)))(((((....)))))..)))).))))))...... ( -37.90) >consensus UUCGC_UUAAC_UUUGCAACGGGGUGUGAACAGUGAUUUUGUUGCUAUUGCACAAAUUGCACUGACUGCGUGGAUCUUGUUGU_________UGUUGCAGUUGCCACUGCAGUUGCUAAA ...............(((((((((((....(((((..(((((.((....)))))))...)))))((((((.........................))))))...)))))).))))).... (-28.47 = -29.03 + 0.56)

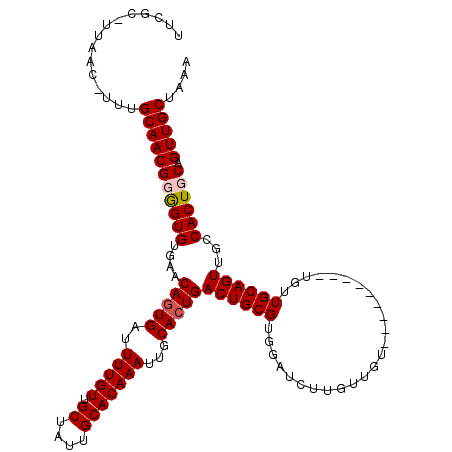
| Location | 6,743,110 – 6,743,220 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -24.08 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6743110 110 - 27905053 UUUAGCAACUGCAGUGGCAACUGCAACA---------ACAACAAGAUCCACGCAGUCAGUGCAAUUUGUGCAAUAGCAACAAAAUCACUGUUCACACCCCGUUGCAAA-GUUAAUGCAAA ....(((((((((((....))))))...---------...................(((((...(((((((....)).)))))..)))))..........)))))...-........... ( -25.40) >DroSec_CAF1 7890 109 - 1 UUUAGCAACUGCAGUGGCAACUGCAACA---------ACAACAAGAUCCACGCAGUCAGUGCAAUUUGUGCAAUAGCAACAAAAUCACUGUUCACACCACGUUGCAAA-GUUAA-GCGAA ....(((((((((((....))))))...---------...................(((((...(((((((....)).)))))..)))))..........)))))...-.....-..... ( -25.40) >DroSim_CAF1 12229 109 - 1 UUUAGCAACUGCAGUGGCAACUGCAACA---------ACAACAAGAUCCACGCAGUCAGUGCAAUUUGUGCAAUAGCAACAAAAUCACUGUUCACACCACGUUGCAAA-GUUAA-GCGAA ....(((((((((((....))))))...---------...................(((((...(((((((....)).)))))..)))))..........)))))...-.....-..... ( -25.40) >DroYak_CAF1 3209 117 - 1 UUUAGCAACUGCAGUGGCAACUGCAACAGCAGCAACAAAAACAAGUUCCACGCAGUCAUUGCAAUUUGUGCAACAGCAACAAAAUCACUGUUCACACUCCGUUGCAAAAGUU---UCAAA ....((((((((((((((..((((....)))).(((........))).......)))))))))...((((.(((((...........)))))))))....))))).......---..... ( -31.10) >consensus UUUAGCAACUGCAGUGGCAACUGCAACA_________ACAACAAGAUCCACGCAGUCAGUGCAAUUUGUGCAAUAGCAACAAAAUCACUGUUCACACCACGUUGCAAA_GUUAA_GCAAA ....(((((((((((....))))))...............................(((((...(((((((....)).)))))..)))))..........)))))............... (-24.08 = -24.32 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:13 2006