
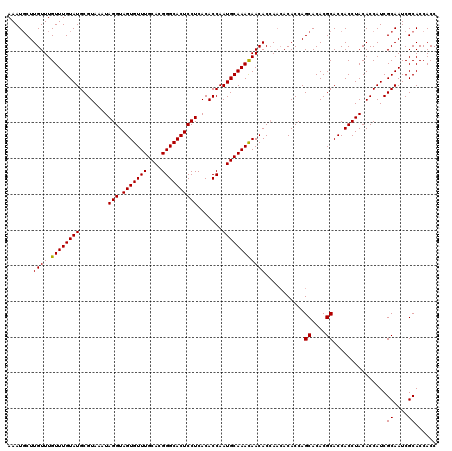
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,740,065 – 6,740,343 |
| Length | 278 |
| Max. P | 0.816716 |

| Location | 6,740,065 – 6,740,183 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6740065 118 + 27905053 AAAUGCUUGUUUGUUUGUAUGCGUAAAUAGGUAGUGUUUGCACGGGCACUCCUCACACCAAUGCAAAUAACACCA--ACACCAGCACACGCACCACCUACACCAUCGCAAUCGCACCACC ...((((((((.((((((.(((((....(((.(((((((....)))))))))).......))))).)))).)).)--)))..))))....................((....))...... ( -22.70) >DroSec_CAF1 4872 120 + 1 AAAUUCUUGUUUGUUUGUAUGCGUAAAUAGGUAGUGUUUGCACGGGCACUCCUCACACCAAUGCAAACAACACCAACACACCAGCACACGCACCACCUACACCAUCGCAAUCGCACCACC ...........(((..((.(((((.....(((..((((((((..((...........))..))))))))..)))..(......)...)))))..))..))).....((....))...... ( -23.20) >DroSim_CAF1 9203 119 + 1 AAAUGCUUGUUUGUUUGUAUGCGUAAAUAGGUAGUGUUUGCACGGGCACUCCUCACACCAAUGCAAACAACAC-AACACACCAGCACACGCACCACCUACACCAUCGCAAUCGCACCACC ...(((((((((((((((((........(((.(((((((....)))))))))).......)))))))))).))-)).......((....))...............)))........... ( -26.06) >consensus AAAUGCUUGUUUGUUUGUAUGCGUAAAUAGGUAGUGUUUGCACGGGCACUCCUCACACCAAUGCAAACAACACCAACACACCAGCACACGCACCACCUACACCAUCGCAAUCGCACCACC .......((((.((((((((........(((.(((((((....)))))))))).......))))))))))))...........((....))...............((....))...... (-22.25 = -22.03 + -0.22)


| Location | 6,740,143 – 6,740,263 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -24.37 |
| Energy contribution | -24.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6740143 120 + 27905053 CCAGCACACGCACCACCUACACCAUCGCAAUCGCACCACCGACUGGCACUUGAAUAAACAUAAAUGCAAGACGUUUAUGUUAAAAGCACCCAACGGGGGCACUUAUCAAGCACCGAACGU ...((....))...............((....)).....((..(((..(((((...((((((((((.....))))))))))....((.(((....))))).....)))))..)))..)). ( -26.40) >DroSec_CAF1 4952 120 + 1 CCAGCACACGCACCACCUACACCAUCGCAAUCGCACCACCGACUGACACUUGAAUAAACAUAAAUGCAAGACGUUUAUGUUAAAAGCACCCAACGGGGGCACUUAUCAAGCACCGAACGU ...((....)).............(((((.(((......))).))...(((((...((((((((((.....))))))))))....((.(((....))))).....)))))...))).... ( -24.30) >DroSim_CAF1 9282 120 + 1 CCAGCACACGCACCACCUACACCAUCGCAAUCGCACCACCGACUGACACUUGAAUAAACAUAAAUGCAAGACGUUUAUGUUAAAAGCACCCAACGGGGGCACUUAUCAAGCACCGAACGU ...((....)).............(((((.(((......))).))...(((((...((((((((((.....))))))))))....((.(((....))))).....)))))...))).... ( -24.30) >consensus CCAGCACACGCACCACCUACACCAUCGCAAUCGCACCACCGACUGACACUUGAAUAAACAUAAAUGCAAGACGUUUAUGUUAAAAGCACCCAACGGGGGCACUUAUCAAGCACCGAACGU ...((....)).............(((((.(((......))).))...(((((...((((((((((.....))))))))))....((.(((....))))).....)))))...))).... (-24.37 = -24.37 + -0.00)

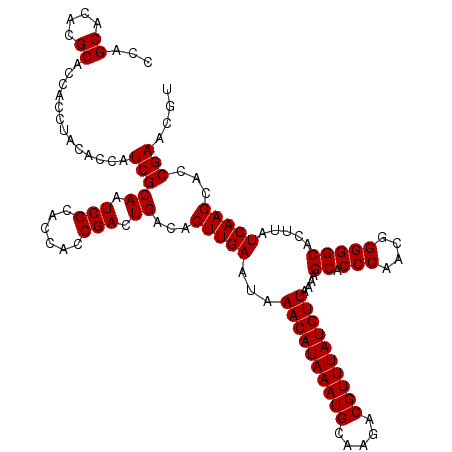

| Location | 6,740,223 – 6,740,343 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.57 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -36.75 |
| Energy contribution | -36.87 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6740223 120 - 27905053 GGAGGGGGAGUGUCAGUGGCAGUGGGCGUGGCAGGAUAAGGGAGCGGGCUCUGAAGUUAGAAGUGAAUCCUUAGCUGACAACGUUCGGUGCUUGAUAAGUGCCCCCGUUGGGUGCUUUUA ...(((((..(((((..((((.(((((((..(((..((((((.((.((((....))))....))...)))))).)))...))))))).)))))))))....))))).............. ( -43.50) >DroSec_CAF1 5032 110 - 1 GG----GG------AGUGGCAGUGGGCGUGGCAGGAUAAGGGAGCGGGCUCUGAAGUUAGAACUGAAUCCUUAGCUGACAACGUUCGGUGCUUGAUAAGUGCCCCCGUUGGGUGCUUUUA ..----..------...((((.(((((((..(((..((((((..(((..((((....)))).)))..)))))).)))...))))))).))))....(((..(((.....)))..)))... ( -40.30) >DroSim_CAF1 9362 110 - 1 GG----GG------AGUGGCAGUGGGCGUGGCAGGAUAAGGGAACGGGCUCUGAAGUUAGAACUGAAUCUUUAGCUGACAACGUUCGGUGCUUGAUAAGUGCCCCCGUUGGGUGCUUUUA ..----..------...((((.(((((((..(((..((((((..(((..((((....)))).)))..)))))).)))...))))))).))))....(((..(((.....)))..)))... ( -36.20) >consensus GG____GG______AGUGGCAGUGGGCGUGGCAGGAUAAGGGAGCGGGCUCUGAAGUUAGAACUGAAUCCUUAGCUGACAACGUUCGGUGCUUGAUAAGUGCCCCCGUUGGGUGCUUUUA .................((((.(((((((..(((..((((((..(((..((((....)))).)))..)))))).)))...))))))).))))....(((..(((.....)))..)))... (-36.75 = -36.87 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:04 2006