
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,738,590 – 6,738,910 |
| Length | 320 |
| Max. P | 0.973886 |

| Location | 6,738,590 – 6,738,710 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -29.03 |
| Energy contribution | -29.37 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6738590 120 - 27905053 CGUGGCUGGAAUAGAGAUUGGCUAAAAGCAUGCCAACAGCAUCUGGGCCAAACGCCAGCGCACAAAAAUUAAUAUGCAAUUAAAUGAAAAUUAGCAAAAAUGUGUGGCGAAUUGAUAAUA .((((((((......(.((((((...((.((((.....)))))).)))))).).))))).)))....((((((.(((.....(((....))).(((......))).))).)))))).... ( -31.60) >DroSec_CAF1 3434 120 - 1 CGUGGCUGGAAUAGAGAUUGACUAAAAGCAUGCCAACAGCAUCUGGGCCAAACGCCAGCGCACAAAAAUUAAUAUGCAAUUAAAUGAAAAUUAGCAAAAAUGUGUGGCGAAUUGAUAAUA .((((((((......(.(((.((...((.((((.....)))))).)).))).).))))).)))....((((((.(((.....(((....))).(((......))).))).)))))).... ( -24.50) >DroSim_CAF1 7790 120 - 1 CGUGGCUGGAAUAGAGAUUGGCUAAAAGCAUGCCAACAGCAUCUGGGCCAAACGCCAGCGCACAAAAAUUAAUAUGCAAUUAAAUGAAAAUUAGCAAAAAUGUGUGGCGAAUUGAUAAUA .((((((((......(.((((((...((.((((.....)))))).)))))).).))))).)))....((((((.(((.....(((....))).(((......))).))).)))))).... ( -31.60) >consensus CGUGGCUGGAAUAGAGAUUGGCUAAAAGCAUGCCAACAGCAUCUGGGCCAAACGCCAGCGCACAAAAAUUAAUAUGCAAUUAAAUGAAAAUUAGCAAAAAUGUGUGGCGAAUUGAUAAUA .((((((((......(.((((((...((.((((.....)))))).)))))).).))))).)))....((((((.(((.....(((....))).(((......))).))).)))))).... (-29.03 = -29.37 + 0.33)



| Location | 6,738,630 – 6,738,750 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -28.26 |
| Energy contribution | -28.04 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6738630 120 + 27905053 AUUGCAUAUUAAUUUUUGUGCGCUGGCGUUUGGCCCAGAUGCUGUUGGCAUGCUUUUAGCCAAUCUCUAUUCCAGCCACGCCACCACUCUCACGGCCACACUUUUCACUUGCCACUUGCC ...(((((........)))))((((((((..(((...((((..((((((.........))))))...))))...)))))))))..........(((..............)))....)). ( -28.94) >DroSec_CAF1 3474 120 + 1 AUUGCAUAUUAAUUUUUGUGCGCUGGCGUUUGGCCCAGAUGCUGUUGGCAUGCUUUUAGUCAAUCUCUAUUCCAGCCACGCCACCACUCUCACGGCCACACUUUUCACUUGCCACUUGCC ...(((((........)))))((((((((.(((((.(((.(.((.((((..(((..(((.......)))....)))...)))).))))))...))))).)).........))))...)). ( -28.60) >DroSim_CAF1 7830 120 + 1 AUUGCAUAUUAAUUUUUGUGCGCUGGCGUUUGGCCCAGAUGCUGUUGGCAUGCUUUUAGCCAAUCUCUAUUCCAGCCACGCCACCACUCUCACGGCCACACUUUUCACUUGCCACUUGCC ...(((((........)))))((((((((..(((...((((..((((((.........))))))...))))...)))))))))..........(((..............)))....)). ( -28.94) >consensus AUUGCAUAUUAAUUUUUGUGCGCUGGCGUUUGGCCCAGAUGCUGUUGGCAUGCUUUUAGCCAAUCUCUAUUCCAGCCACGCCACCACUCUCACGGCCACACUUUUCACUUGCCACUUGCC ...(((((........)))))((((((((..(((...((((..((((((.........))))))...))))...)))))))))..........(((..............)))....)). (-28.26 = -28.04 + -0.22)


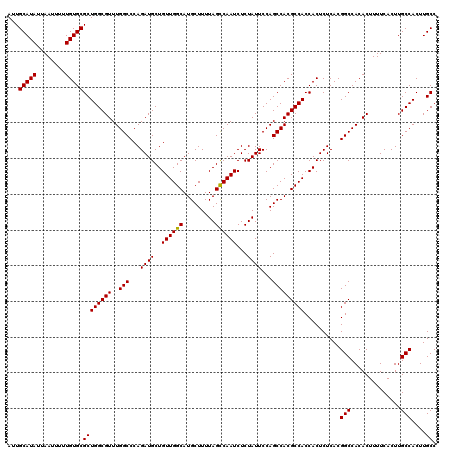
| Location | 6,738,630 – 6,738,750 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -35.43 |
| Energy contribution | -35.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6738630 120 - 27905053 GGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGGCGUGGCUGGAAUAGAGAUUGGCUAAAAGCAUGCCAACAGCAUCUGGGCCAAACGCCAGCGCACAAAAAUUAAUAUGCAAU .(((...(((.(.(....).)...)))......(((.((((((...(((..(((((.(((((.........)))))...).))))..))).)))))).))).............)))... ( -38.00) >DroSec_CAF1 3474 120 - 1 GGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGGCGUGGCUGGAAUAGAGAUUGACUAAAAGCAUGCCAACAGCAUCUGGGCCAAACGCCAGCGCACAAAAAUUAAUAUGCAAU .(((...(((.(.(....).)...)))......(((.((((((((((....(((.......)))..((.((((.....)))))).))))..)))))).))).............)))... ( -32.20) >DroSim_CAF1 7830 120 - 1 GGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGGCGUGGCUGGAAUAGAGAUUGGCUAAAAGCAUGCCAACAGCAUCUGGGCCAAACGCCAGCGCACAAAAAUUAAUAUGCAAU .(((...(((.(.(....).)...)))......(((.((((((...(((..(((((.(((((.........)))))...).))))..))).)))))).))).............)))... ( -38.00) >consensus GGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGGCGUGGCUGGAAUAGAGAUUGGCUAAAAGCAUGCCAACAGCAUCUGGGCCAAACGCCAGCGCACAAAAAUUAAUAUGCAAU .(((...(((.(.(....).)...)))......(((.((((((...(((..(((((.(((((.........)))))...).))))..))).)))))).))).............)))... (-35.43 = -35.77 + 0.33)

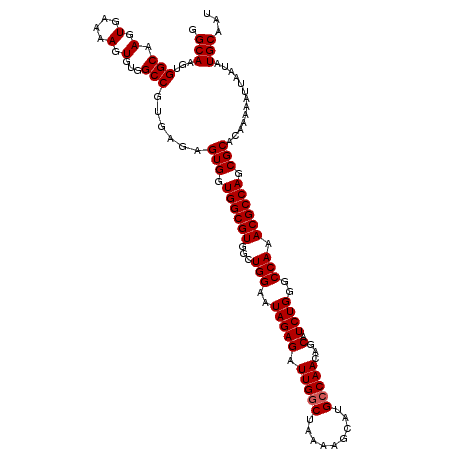

| Location | 6,738,670 – 6,738,790 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -35.33 |
| Energy contribution | -35.33 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.99 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6738670 120 - 27905053 GGCACUGAAAGCAAAAGUGUUUUACUGGACUCUCUCGAGUGGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGGCGUGGCUGGAAUAGAGAUUGGCUAAAAGCAUGCCAACAGC ((((((.........))))))...(((.((((((.((.((.(((..(..(....)..)..))).)))).))))))((((((.((((..(........)..))))...)).))))..))). ( -35.90) >DroSec_CAF1 3514 120 - 1 GGCACUGAAAGCAAAAGUGUUUUACUGGACUCUCUCGAGUGGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGGCGUGGCUGGAAUAGAGAUUGACUAAAAGCAUGCCAACAGC ((((((.........))))))...(((.((((((.((.((.(((..(..(....)..)..))).)))).))))))..(((((((.((....(((.......)))..))))))))).))). ( -35.20) >DroSim_CAF1 7870 120 - 1 GGCACUGAAAGCAAAAGUGUUUUACUGGACUCUCUCGAGUGGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGGCGUGGCUGGAAUAGAGAUUGGCUAAAAGCAUGCCAACAGC ((((((.........))))))...(((.((((((.((.((.(((..(..(....)..)..))).)))).))))))((((((.((((..(........)..))))...)).))))..))). ( -35.90) >consensus GGCACUGAAAGCAAAAGUGUUUUACUGGACUCUCUCGAGUGGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGGCGUGGCUGGAAUAGAGAUUGGCUAAAAGCAUGCCAACAGC ((((((.........))))))...(((.((((((.((.((.(((..(..(....)..)..))).)))).))))))..(((((((.((....(((.......)))..))))))))).))). (-35.33 = -35.33 + 0.00)

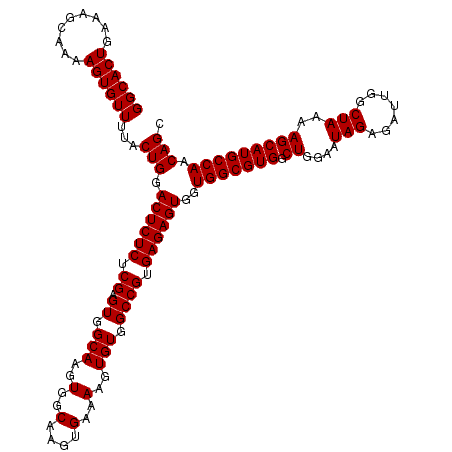
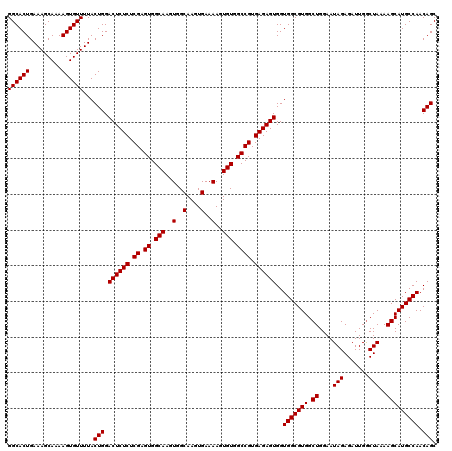
| Location | 6,738,710 – 6,738,830 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -36.90 |
| Energy contribution | -36.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6738710 120 + 27905053 CCACCACUCUCACGGCCACACUUUUCACUUGCCACUUGCCACUCGAGAGAGUCCAGUAAAACACUUUUGCUUUCAGUGCCUGGCAGGCAGAAGUUUCCGCUGCCGGUGAUUAAUGUGCCG ............((((.(((.(..((((((((((...(((((((....))))..((((((.....))))))....).)).)))))(((((.........))))))))))..).))))))) ( -36.90) >DroSec_CAF1 3554 120 + 1 CCACCACUCUCACGGCCACACUUUUCACUUGCCACUUGCCACUCGAGAGAGUCCAGUAAAACACUUUUGCUUUCAGUGCCUGGCAGGCAGAAGUUUCCGCUGCCGGUGAUUAAUGUGCCG ............((((.(((.(..((((((((((...(((((((....))))..((((((.....))))))....).)).)))))(((((.........))))))))))..).))))))) ( -36.90) >DroSim_CAF1 7910 120 + 1 CCACCACUCUCACGGCCACACUUUUCACUUGCCACUUGCCACUCGAGAGAGUCCAGUAAAACACUUUUGCUUUCAGUGCCUGGCAGGCAGAAGUUUCCGCUGCCGGUGAUUAAUGUGCCG ............((((.(((.(..((((((((((...(((((((....))))..((((((.....))))))....).)).)))))(((((.........))))))))))..).))))))) ( -36.90) >consensus CCACCACUCUCACGGCCACACUUUUCACUUGCCACUUGCCACUCGAGAGAGUCCAGUAAAACACUUUUGCUUUCAGUGCCUGGCAGGCAGAAGUUUCCGCUGCCGGUGAUUAAUGUGCCG ............((((.(((.(..((((((((((...(((((((....))))..((((((.....))))))....).)).)))))(((((.........))))))))))..).))))))) (-36.90 = -36.90 + -0.00)

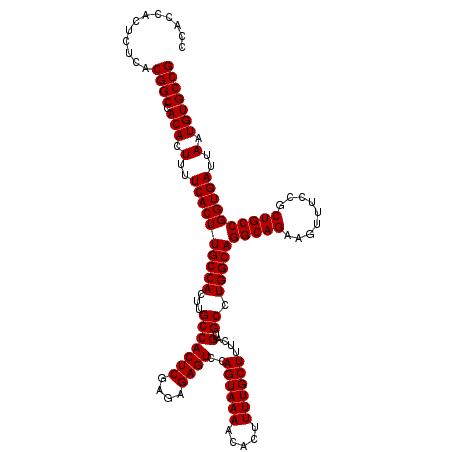
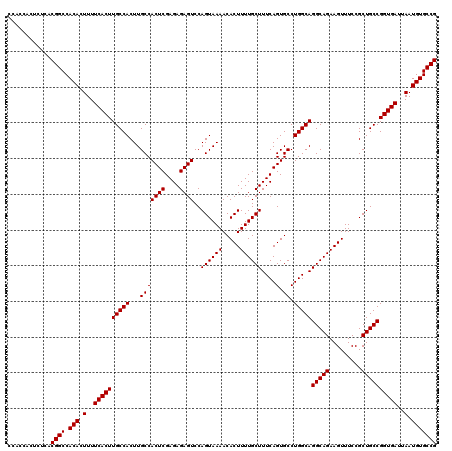
| Location | 6,738,710 – 6,738,830 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -42.00 |
| Energy contribution | -42.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6738710 120 - 27905053 CGGCACAUUAAUCACCGGCAGCGGAAACUUCUGCCUGCCAGGCACUGAAAGCAAAAGUGUUUUACUGGACUCUCUCGAGUGGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGG ............(((((((((.(....)..)))))..(((((((((.........)))))....))))((((((.((.((.(((..(..(....)..)..))).)))).)))))))))). ( -42.00) >DroSec_CAF1 3554 120 - 1 CGGCACAUUAAUCACCGGCAGCGGAAACUUCUGCCUGCCAGGCACUGAAAGCAAAAGUGUUUUACUGGACUCUCUCGAGUGGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGG ............(((((((((.(....)..)))))..(((((((((.........)))))....))))((((((.((.((.(((..(..(....)..)..))).)))).)))))))))). ( -42.00) >DroSim_CAF1 7910 120 - 1 CGGCACAUUAAUCACCGGCAGCGGAAACUUCUGCCUGCCAGGCACUGAAAGCAAAAGUGUUUUACUGGACUCUCUCGAGUGGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGG ............(((((((((.(....)..)))))..(((((((((.........)))))....))))((((((.((.((.(((..(..(....)..)..))).)))).)))))))))). ( -42.00) >consensus CGGCACAUUAAUCACCGGCAGCGGAAACUUCUGCCUGCCAGGCACUGAAAGCAAAAGUGUUUUACUGGACUCUCUCGAGUGGCAAGUGGCAAGUGAAAAGUGUGGCCGUGAGAGUGGUGG ............(((((((((.(....)..)))))..(((((((((.........)))))....))))((((((.((.((.(((..(..(....)..)..))).)))).)))))))))). (-42.00 = -42.00 + 0.00)

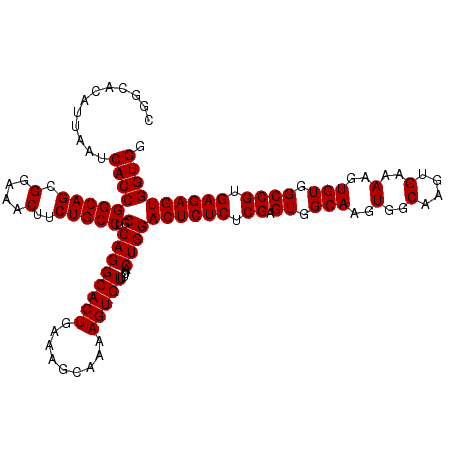
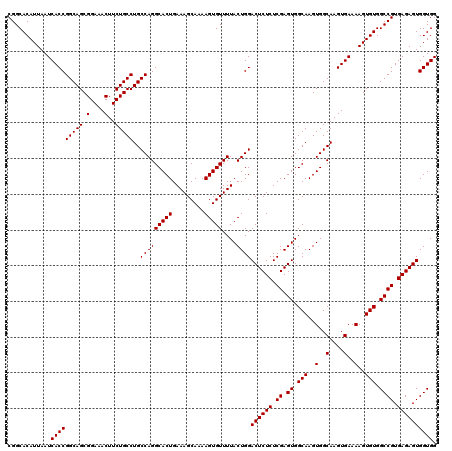
| Location | 6,738,790 – 6,738,910 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -44.19 |
| Consensus MFE | -42.92 |
| Energy contribution | -43.26 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6738790 120 - 27905053 GAGCUGACUAGAUGGCUACCUGGGUAGAUGGCCAAGCCGUAUCUCUUGGCCAUGGUUUAAAGCUGGCCCGAACAUUAUUGCGGCACAUUAAUCACCGGCAGCGGAAACUUCUGCCUGCCA .((((...(((((..((((....))))(((((((((........))))))))).))))).)))).((((((......))).)))............(((((((((....)))).))))). ( -42.40) >DroSec_CAF1 3634 120 - 1 GAGCUGACUAGAUGGCUAGCUGGGUAGCUGGCCAAGCCGUAUCUCUUGGCCAUGGUUUAAAGCUGGCCCGAACAUUAUUGCGGCACAUUAAUCACCGGCAGCGGAAACUUCUGCCUGCCA ..(.(((.(((.(((((((((....))))))))).((((((......(((((..((.....)))))))..........))))))...))).))).)(((((((((....)))).))))). ( -45.09) >DroSim_CAF1 7990 120 - 1 GAGCUGACUAGAUGGCUAGCUGGGUAGCUGGCCAAGCCGUAUCUCUUGGCCAUGGUUUAAAGCUGGCCCGAACAUUAUUGCGGCACAUUAAUCACCGGCAGCGGAAACUUCUGCCUGCCA ..(.(((.(((.(((((((((....))))))))).((((((......(((((..((.....)))))))..........))))))...))).))).)(((((((((....)))).))))). ( -45.09) >consensus GAGCUGACUAGAUGGCUAGCUGGGUAGCUGGCCAAGCCGUAUCUCUUGGCCAUGGUUUAAAGCUGGCCCGAACAUUAUUGCGGCACAUUAAUCACCGGCAGCGGAAACUUCUGCCUGCCA ..(.(((.(((.(((((((((....))))))))).((((((......(((((..((.....)))))))..........))))))...))).))).)(((((((((....)))).))))). (-42.92 = -43.26 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:59 2006