
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,736,771 – 6,737,006 |
| Length | 235 |
| Max. P | 0.977808 |

| Location | 6,736,771 – 6,736,891 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -30.02 |
| Energy contribution | -29.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6736771 120 - 27905053 AAUCAAAAGUGUUUCGCCAUCGAGGCGUAGCACACAUUUAUGCAUAAAUUGUAUUUACAUUUCUAUAUCCAGCCAGAAAAAAGUGGCAUAUCGCCGCGCAAUGUCCGCUGCUUGUUGCAC ........(((((.((((.....)))).))))).......((((.....(((....))).....(((..((((..((.....(((((.....)))))......)).))))..))))))). ( -30.40) >DroSec_CAF1 1689 120 - 1 AAUCAAAAGUGUUUCGCCAUCGAGGCGUAGCACACAUUUAUGCAUAAAUUGUAUUUACAUUCCUAUAUCCAGCCAGAAAAAAGUGGCAUAUCGCCGCGCUUUGUCCGCUGCUUGUUGCAC ........(((((.((((.....)))).))))).......((((.....(((....))).....(((..((((....(((..(((((.....)))))..)))....))))..))))))). ( -30.80) >DroSim_CAF1 5964 119 - 1 AAUCAAAAGUGUUUCGCCAUCGAGGCGUAGCACACAAUUAUGCAUAAAUUGUAUUUACAUUUCUAUAUCCAGUCAGAA-AAAGUGGCUUAUCGCCGCGCAAUGUCCGCUGCUUGUUGCAC ........(((((.((((.....)))).))))).((((...(((...(((((.......(((((..........))))-)..(((((.....))))))))))......)))..))))... ( -29.70) >consensus AAUCAAAAGUGUUUCGCCAUCGAGGCGUAGCACACAUUUAUGCAUAAAUUGUAUUUACAUUUCUAUAUCCAGCCAGAAAAAAGUGGCAUAUCGCCGCGCAAUGUCCGCUGCUUGUUGCAC ........(((((.((((.....)))).))))).......((((.....(((....))).....(((..((((..((.....(((((.....)))))......)).))))..))))))). (-30.02 = -29.80 + -0.22)


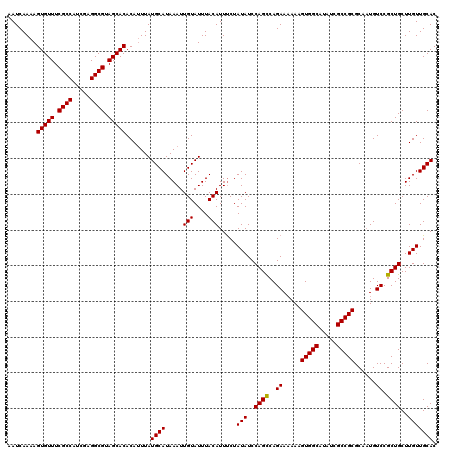
| Location | 6,736,851 – 6,736,966 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -18.10 |
| Energy contribution | -19.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6736851 115 + 27905053 UAAAUGUGUGCUACGCCUCGAUGGCGAAACACUUUUGAUUUACAACACUGUGUGCAAAAAGUAUUCG-----CAUUGCCAUGCCCAUGCUUUUUGCUCAGAUCUUUCAAUUUCAAUGUCA ((((.((((....((((.....))))..)))).))))..........(((...(((((((((((..(-----(((....))))..))))))))))).))).................... ( -30.60) >DroSec_CAF1 1769 102 + 1 UAAAUGUGUGCUACGCCUCGAUGGCGAAACACUUUUGAUUUACAACACUGUGUGCAAAAAGUAUUCGGAUGGCAUGGCAAUGCCA--A----------------UGACAUUUCAAUGUCA ((((.((((....((((.....))))..)))).))))..........(((.((((.....)))).))).((((((....))))))--.----------------((((((....)))))) ( -29.00) >DroSim_CAF1 6043 118 + 1 UAAUUGUGUGCUACGCCUCGAUGGCGAAACACUUUUGAUUUACAACACUGUGUGCAAAAAGUAUUCGGAUGGCAUGGCAAUGCCC--GCUUUUUGCUCAGAUCUUUCGAUUUCAAUGUCA (((..((((....((((.....))))..))))..)))..........(((...(((((((((........(((((....))))).--))))))))).))).......(((......))). ( -30.50) >consensus UAAAUGUGUGCUACGCCUCGAUGGCGAAACACUUUUGAUUUACAACACUGUGUGCAAAAAGUAUUCGGAUGGCAUGGCAAUGCCC__GCUUUUUGCUCAGAUCUUUCAAUUUCAAUGUCA .....((((....((((.....))))..))))..((((.........(((.((((.....)))).)))..(((((....)))))...........................))))..... (-18.10 = -19.10 + 1.00)


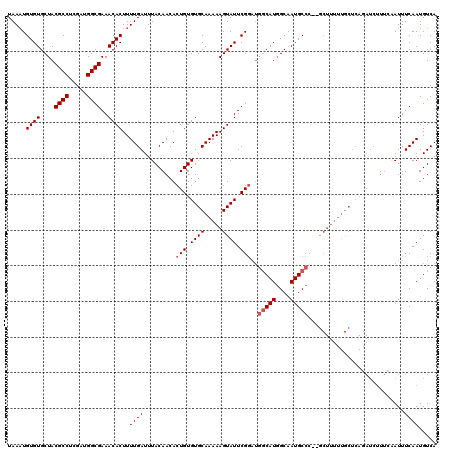
| Location | 6,736,851 – 6,736,966 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -22.90 |
| Energy contribution | -23.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6736851 115 - 27905053 UGACAUUGAAAUUGAAAGAUCUGAGCAAAAAGCAUGGGCAUGGCAAUG-----CGAAUACUUUUUGCACACAGUGUUGUAAAUCAAAAGUGUUUCGCCAUCGAGGCGUAGCACACAUUUA .(((((((..(((....)))....((((((((.((..((((....)))-----)..)).))))))))...)))))))...........(((((.((((.....)))).)))))....... ( -34.60) >DroSec_CAF1 1769 102 - 1 UGACAUUGAAAUGUCA----------------U--UGGCAUUGCCAUGCCAUCCGAAUACUUUUUGCACACAGUGUUGUAAAUCAAAAGUGUUUCGCCAUCGAGGCGUAGCACACAUUUA ((((((....))))))----------------.--((((((....))))))...........((((((........))))))......(((((.((((.....)))).)))))....... ( -27.00) >DroSim_CAF1 6043 118 - 1 UGACAUUGAAAUCGAAAGAUCUGAGCAAAAAGC--GGGCAUUGCCAUGCCAUCCGAAUACUUUUUGCACACAGUGUUGUAAAUCAAAAGUGUUUCGCCAUCGAGGCGUAGCACACAAUUA .(((((((..(((....)))....(((((((((--((((((....))))...)))....))))))))...)))))))...........(((((.((((.....)))).)))))....... ( -36.40) >consensus UGACAUUGAAAUCGAAAGAUCUGAGCAAAAAGC__GGGCAUUGCCAUGCCAUCCGAAUACUUUUUGCACACAGUGUUGUAAAUCAAAAGUGUUUCGCCAUCGAGGCGUAGCACACAUUUA .(((((((............................(((((....)))))...((((.....))))....)))))))...........(((((.((((.....)))).)))))....... (-22.90 = -23.57 + 0.67)


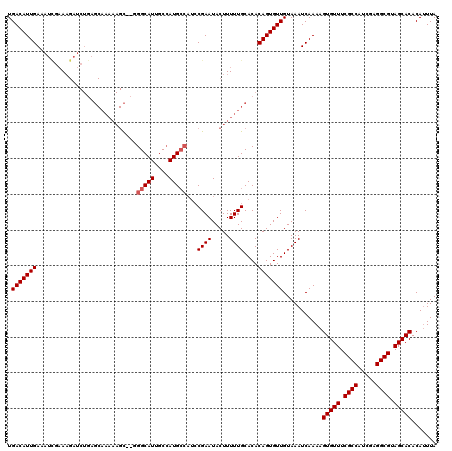
| Location | 6,736,891 – 6,737,006 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -16.30 |
| Energy contribution | -17.63 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6736891 115 - 27905053 AUAAGCUGCAAAAAUGCAAUAUUUUUUUUCGCUACCAAAAUGACAUUGAAAUUGAAAGAUCUGAGCAAAAAGCAUGGGCAUGGCAAUG-----CGAAUACUUUUUGCACACAGUGUUGUA ......((((....)))).....................(..((((((..(((....)))....((((((((.((..((((....)))-----)..)).))))))))...))))))..). ( -25.40) >DroSec_CAF1 1809 101 - 1 AUAAGCUGCAAAAAUGCAAUAUUU-UUUGCGCUACCAAAAUGACAUUGAAAUGUCA----------------U--UGGCAUUGCCAUGCCAUCCGAAUACUUUUUGCACACAGUGUUGUA ...(((.(((((((........))-)))))))).....((((((((....))))))----------------)--)(((((....))))).....((((((..........))))))... ( -24.20) >DroSim_CAF1 6083 118 - 1 AUAAGCUGCAAAAAUGCAAUAUUUUUUUGCGCUACCAAAAUGACAUUGAAAUCGAAAGAUCUGAGCAAAAAGC--GGGCAUUGCCAUGCCAUCCGAAUACUUUUUGCACACAGUGUUGUA ...(((.(((((((.........))))))))))......(..((((((..(((....)))....(((((((((--((((((....))))...)))....))))))))...))))))..). ( -32.90) >consensus AUAAGCUGCAAAAAUGCAAUAUUUUUUUGCGCUACCAAAAUGACAUUGAAAUCGAAAGAUCUGAGCAAAAAGC__GGGCAUUGCCAUGCCAUCCGAAUACUUUUUGCACACAGUGUUGUA ...(((.(((((((.........))))))))))......(..((((((............................(((((....)))))...((((.....))))....))))))..). (-16.30 = -17.63 + 1.33)

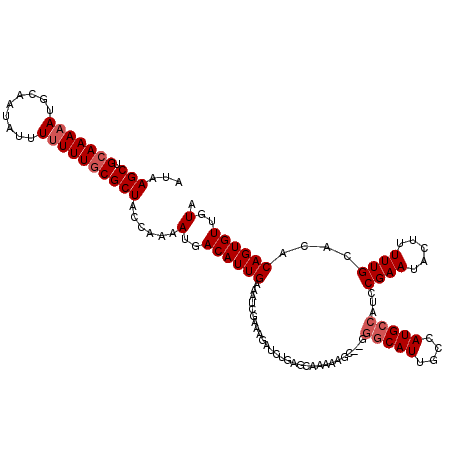

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:49 2006