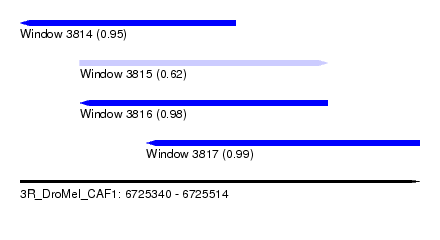
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,725,340 – 6,725,514 |
| Length | 174 |
| Max. P | 0.994829 |

| Location | 6,725,340 – 6,725,434 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 92.20 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -21.28 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6725340 94 - 27905053 UUCACACUUAAAUGAACUUAGCCCGGUUCAUAACAGGCCAUAUUAAUUGGCCAACAGCCAGAUGAUUCGGCUGCUUAAGCACUCCUCUCAUAAA ......((((((((((((......)))))))....(((((.......)))))..(((((.((....))))))).)))))............... ( -25.00) >DroEre_CAF1 9956 94 - 1 UUCACACUUAAAUGGACUUAGCUCGGUCCAUAACUGGCCAUAUUAAUUGGCAAAGAGCCAGAUGAUUCGGCUGCUUAAGCUCUCCUCGCAUAAA ......((((((((((((......)))))))....((((..((((.(((((.....))))).))))..))))..)))))............... ( -25.00) >DroYak_CAF1 9920 94 - 1 UUCACACUUAAAUGGACUUAGCUCGCUCCAUAACAGGCCAUAUUAAUUGGCAAAGAGCCAGUUGAUUCGGCUGCUUAAACUCUCCUCUCAUAAA ...........((((((.......).)))))....((((..((((((((((.....))))))))))..))))...................... ( -22.10) >consensus UUCACACUUAAAUGGACUUAGCUCGGUCCAUAACAGGCCAUAUUAAUUGGCAAAGAGCCAGAUGAUUCGGCUGCUUAAGCUCUCCUCUCAUAAA ......((((((((((((......)))))))....((((..((((.(((((.....))))).))))..))))..)))))............... (-21.28 = -21.50 + 0.22)



| Location | 6,725,366 – 6,725,474 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -21.24 |
| Energy contribution | -22.17 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6725366 108 + 27905053 GAAUCAUCUGGCUGUUGGCCAAUUAAUAUGGCCUGUUAUGAACCG-GGCUAAGUUCAUUUAAGUGUGAAAUACUUAAAAAGCAUUCGACGGCCUGUGGCUAUAUGUACA ....(((.((((..(.((((........(((((((........))-))))).(((..(((((((((...))))))))).))).......)))).)..)))).))).... ( -33.30) >DroSec_CAF1 8574 109 + 1 GAAUCAUCUGGCUGUUGGCCAAUUAAUAUGGCCUGUUAUGAACUAUGGCUAAGUUCAUUUAAGUGUGAAAUACAUAAAAAGCAUUCGAUGGCCUGUGGCUCUGUGUACA ((..(((..((((((((((((.......)))))(((((((((((.......)))))))....(((((...)))))....))))...))))))).)))..))........ ( -28.10) >DroEre_CAF1 9982 104 + 1 GAAUCAUCUGGCUCUUUGCCAAUUAAUAUGGCCAGUUAUGGACCG-AGCUAAGUCCAUUUAAGUGUGAAAUACUUAAACAGCAUUCGAUGGCCUGUGGCUAU----ACA ........((((.....)))).....(((((((....((((((..-......))))))((((((((...))))))))(((((........).))))))))))----).. ( -27.50) >DroYak_CAF1 9946 104 + 1 GAAUCAACUGGCUCUUUGCCAAUUAAUAUGGCCUGUUAUGGAGCG-AGCUAAGUCCAUUUAAGUGUGAAAUACUUAAAAAGCAAUCGAUGGCCUGUGGCUAU----GCA ........((((.....)))).....(((((((.(((((.(((((-(......))..(((((((((...)))))))))..))..)).)))))....))))))----).. ( -24.50) >consensus GAAUCAUCUGGCUCUUGGCCAAUUAAUAUGGCCUGUUAUGAACCG_AGCUAAGUCCAUUUAAGUGUGAAAUACUUAAAAAGCAUUCGAUGGCCUGUGGCUAU____ACA ...(((.(.((((((((((((.......))))).(((.(((((.........)))))(((((((((...))))))))).)))....))))))).))))........... (-21.24 = -22.17 + 0.94)

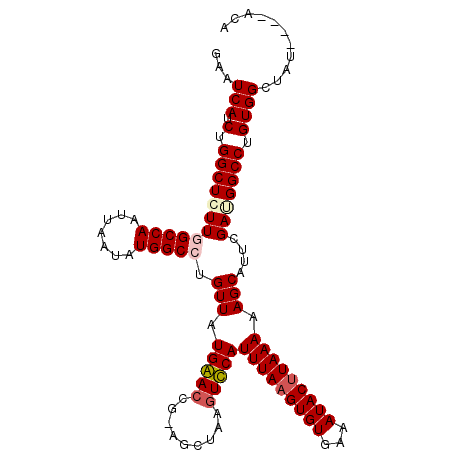
| Location | 6,725,366 – 6,725,474 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -24.11 |
| Energy contribution | -24.17 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6725366 108 - 27905053 UGUACAUAUAGCCACAGGCCGUCGAAUGCUUUUUAAGUAUUUCACACUUAAAUGAACUUAGCC-CGGUUCAUAACAGGCCAUAUUAAUUGGCCAACAGCCAGAUGAUUC ................((((((.(((((((.....))))))).))......(((((((.....-.)))))))....))))..((((.(((((.....))))).)))).. ( -27.60) >DroSec_CAF1 8574 109 - 1 UGUACACAGAGCCACAGGCCAUCGAAUGCUUUUUAUGUAUUUCACACUUAAAUGAACUUAGCCAUAGUUCAUAACAGGCCAUAUUAAUUGGCCAACAGCCAGAUGAUUC ................((((...((((((.......)))))).........(((((((.......)))))))....))))..((((.(((((.....))))).)))).. ( -24.50) >DroEre_CAF1 9982 104 - 1 UGU----AUAGCCACAGGCCAUCGAAUGCUGUUUAAGUAUUUCACACUUAAAUGGACUUAGCU-CGGUCCAUAACUGGCCAUAUUAAUUGGCAAAGAGCCAGAUGAUUC ...----.........(((((..(((((((.....))))))).........(((((((.....-.)))))))...)))))..((((.(((((.....))))).)))).. ( -29.40) >DroYak_CAF1 9946 104 - 1 UGC----AUAGCCACAGGCCAUCGAUUGCUUUUUAAGUAUUUCACACUUAAAUGGACUUAGCU-CGCUCCAUAACAGGCCAUAUUAAUUGGCAAAGAGCCAGUUGAUUC ...----.........((((............((((((.......))))))((((((......-.).)))))....))))..((((((((((.....)))))))))).. ( -25.60) >consensus UGU____AUAGCCACAGGCCAUCGAAUGCUUUUUAAGUAUUUCACACUUAAAUGAACUUAGCC_CGGUCCAUAACAGGCCAUAUUAAUUGGCAAACAGCCAGAUGAUUC ................((((...(((((((.....))))))).........(((((((.......)))))))....))))..((((.(((((.....))))).)))).. (-24.11 = -24.17 + 0.06)

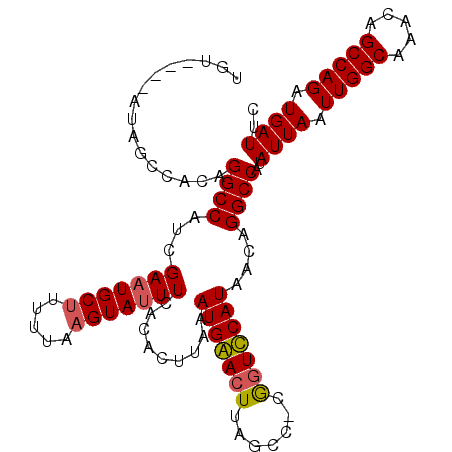

| Location | 6,725,395 – 6,725,514 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -18.88 |
| Energy contribution | -20.25 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6725395 119 - 27905053 CCAAAAAAAAAAAAAAAAACAAACAAACAUGGAGUUAAAAUGUACAUAUAGCCACAGGCCGUCGAAUGCUUUUUAAGUAUUUCACACUUAAAUGAACUUAGCC-CGGUUCAUAACAGGCC .............................((..((((...........))))..))((((((.(((((((.....))))))).))......(((((((.....-.)))))))....)))) ( -21.10) >DroSec_CAF1 8603 116 - 1 CCAUAAAAA----AAAAAACACACACACAUGGAGCUAAAAUGUACACAGAGCCACAGGCCAUCGAAUGCUUUUUAUGUAUUUCACACUUAAAUGAACUUAGCCAUAGUUCAUAACAGGCC .........----................((..(((....((....)).)))..))((((...((((((.......)))))).........(((((((.......)))))))....)))) ( -21.30) >DroEre_CAF1 10011 108 - 1 CCAA---AA----AAAAAAUAUUACCACAUGUGACUAAAAUGU----AUAGCCACAGGCCAUCGAAUGCUGUUUAAGUAUUUCACACUUAAAUGGACUUAGCU-CGGUCCAUAACUGGCC ....---..----................((((.(((......----.))).))))(((((..(((((((.....))))))).........(((((((.....-.)))))))...))))) ( -25.30) >DroYak_CAF1 9975 108 - 1 CCAA---AA----AAAAAUAAUGAACACAUGUGGCUAAAAUGC----AUAGCCACAGGCCAUCGAUUGCUUUUUAAGUAUUUCACACUUAAAUGGACUUAGCU-CGCUCCAUAACAGGCC ....---..----................((((((((......----.))))))))((((............((((((.......))))))((((((......-.).)))))....)))) ( -24.60) >consensus CCAA___AA____AAAAAACAUAAACACAUGGAGCUAAAAUGU____AUAGCCACAGGCCAUCGAAUGCUUUUUAAGUAUUUCACACUUAAAUGAACUUAGCC_CGGUCCAUAACAGGCC .............................((((((((...........))))))))((((...(((((((.....))))))).........(((((((.......)))))))....)))) (-18.88 = -20.25 + 1.38)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:32 2006