
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,725,158 – 6,725,292 |
| Length | 134 |
| Max. P | 0.998220 |

| Location | 6,725,158 – 6,725,278 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.17 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -27.17 |
| Energy contribution | -26.61 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6725158 120 + 27905053 GUUUAUCCACUGGCCGAUUGGCAAAAGAAGCCACAGCAAAAGCUUGCGGUGUGUGUGUGUGUGCAUAUAAUUUGAUAUGCCGGCAAGCUAAAAAGUGGCCAAGGAGGAAAUAGCCAACAA (((..(((..((((((..((((.......)))).......(((((((((((((((((((....)))))......))))))).)))))))......)))))).)))((......))))).. ( -38.00) >DroEre_CAF1 9764 109 + 1 GUUUAUCCACUGGCCGAUUGGCAAAAGAAGCCACAGCAAAAGCUUGCGGUGU--------GUGCAUAUAAUUUGAUAUGCCGGCAAGCUAAAAAGUGGCCAGGAAAGAAAUCCUC---AA .........(((((((..((((.......)))).......((((((((((((--------((.((.......))))))))).)))))))......))))))).............---.. ( -36.20) >DroYak_CAF1 9712 109 + 1 GUUUAUCCACUGGCCGAUUGGCAAAAGAAGCCACAGCAAAAGCUUGCGGUGU--------GUGCAUAUAAUUUGAUAUGCCGGCAAGCUAAAAAGUGGCCAAGAAAGAAAUCCCC---AA ..........((((((..((((.......)))).......((((((((((((--------((.((.......))))))))).)))))))......))))))..............---.. ( -32.70) >DroAna_CAF1 9575 102 + 1 GUC------CGGGUCGAUUGGCAAAAGAAGCCACAGCAAAAGCUUGCGGUGU--------GUGCAUAUAAUUUGAUAUGCCAGUGAGCUAAAAA-UGGCCAAGAAAGAAAUCCGU---AA ...------((((((..(((((......((((((.((((....))))(((((--------((.((.......))))))))).))).))).....-..)))))....))..)))).---.. ( -29.02) >consensus GUUUAUCCACUGGCCGAUUGGCAAAAGAAGCCACAGCAAAAGCUUGCGGUGU________GUGCAUAUAAUUUGAUAUGCCGGCAAGCUAAAAAGUGGCCAAGAAAGAAAUCCCC___AA ...........(((((..((((.......)))).......(((((((.(.............((((((......))))))).)))))))......))))).................... (-27.17 = -26.61 + -0.56)

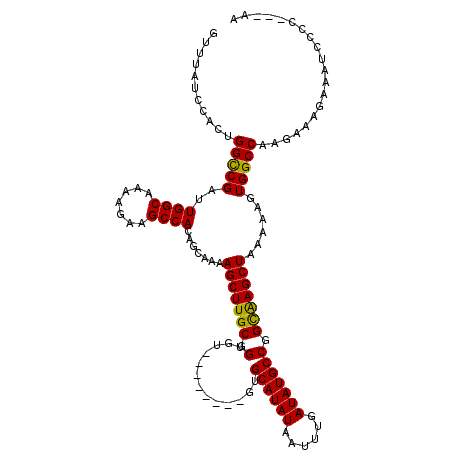

| Location | 6,725,158 – 6,725,278 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.17 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -23.89 |
| Energy contribution | -25.33 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6725158 120 - 27905053 UUGUUGGCUAUUUCCUCCUUGGCCACUUUUUAGCUUGCCGGCAUAUCAAAUUAUAUGCACACACACACACACCGCAAGCUUUUGCUGUGGCUUCUUUUGCCAAUCGGCCAGUGGAUAAAC (..(((((((((........((((((.....(((((((..((((((......))))))...............)))))))......)))))).........))).))))))..)...... ( -32.56) >DroEre_CAF1 9764 109 - 1 UU---GAGGAUUUCUUUCCUGGCCACUUUUUAGCUUGCCGGCAUAUCAAAUUAUAUGCAC--------ACACCGCAAGCUUUUGCUGUGGCUUCUUUUGCCAAUCGGCCAGUGGAUAAAC ..---.((((......))))..(((((....(((((((..((((((......))))))..--------.....)))))))...((((((((.......))))..)))).)))))...... ( -33.30) >DroYak_CAF1 9712 109 - 1 UU---GGGGAUUUCUUUCUUGGCCACUUUUUAGCUUGCCGGCAUAUCAAAUUAUAUGCAC--------ACACCGCAAGCUUUUGCUGUGGCUUCUUUUGCCAAUCGGCCAGUGGAUAAAC (.---.((((.....))))..)(((((....(((((((..((((((......))))))..--------.....)))))))...((((((((.......))))..)))).)))))...... ( -32.50) >DroAna_CAF1 9575 102 - 1 UU---ACGGAUUUCUUUCUUGGCCA-UUUUUAGCUCACUGGCAUAUCAAAUUAUAUGCAC--------ACACCGCAAGCUUUUGCUGUGGCUUCUUUUGCCAAUCGACCCG------GAC ..---.(((...((....(((((..-.....(((......((((((......))))))..--------.(((.((((....)))).))))))......)))))..)).)))------... ( -22.92) >consensus UU___GGGGAUUUCUUUCUUGGCCACUUUUUAGCUUGCCGGCAUAUCAAAUUAUAUGCAC________ACACCGCAAGCUUUUGCUGUGGCUUCUUUUGCCAAUCGGCCAGUGGAUAAAC ...............(((((((((.......(((((((..((((((......))))))...............))))))).......((((.......))))...)))))).)))..... (-23.89 = -25.33 + 1.44)


| Location | 6,725,198 – 6,725,292 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.34 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -15.95 |
| Energy contribution | -15.82 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.62 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6725198 94 + 27905053 AGCUUGCGGUGUGUGUGUGUGUGCAUAUAAUUUGAUAUGCCGGCAAGCUAAAAAGUGGCCAAGGAGGAAAUAGCCAACAACA---CGGCCAAUUU-------CC---------------- (((((((((((((((((((....)))))......))))))).)))))))................((((((.(((.......---.)))..))))-------))---------------- ( -27.70) >DroEre_CAF1 9804 97 + 1 AGCUUGCGGUGU--------GUGCAUAUAAUUUGAUAUGCCGGCAAGCUAAAAAGUGGCCAGGAAAGAAAUCCUC---AAUA---CAGCCAAUUUCCAAUUUCCAAUUUCC--------- ((((((((((((--------((.((.......))))))))).)))))))......((((.((((......)))).---....---..))))....................--------- ( -24.00) >DroYak_CAF1 9752 96 + 1 AGCUUGCGGUGU--------GUGCAUAUAAUUUGAUAUGCCGGCAAGCUAAAAAGUGGCCAAGAAAGAAAUCCCC---AACA---CAGCCAAUUU-------CCAAA---CUGGCCAAUA ((((((((((((--------((.((.......))))))))).)))))))......((((((.....(((((...(---....---..)...))))-------)....---.))))))... ( -28.20) >DroAna_CAF1 9609 78 + 1 AGCUUGCGGUGU--------GUGCAUAUAAUUUGAUAUGCCAGUGAGCUAAAAA-UGGCCAAGAAAGAAAUCCGU---AAGCCACCAGCC------------------------------ .((((((((..(--------(.((((((......)))))))).((.((((....-))))))..........))))---))))........------------------------------ ( -22.30) >consensus AGCUUGCGGUGU________GUGCAUAUAAUUUGAUAUGCCGGCAAGCUAAAAAGUGGCCAAGAAAGAAAUCCCC___AACA___CAGCCAAUUU_______CC________________ (((((((.(.............((((((......))))))).)))))))......((((............................))))............................. (-15.95 = -15.82 + -0.12)



| Location | 6,725,198 – 6,725,292 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.34 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -14.49 |
| Energy contribution | -14.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6725198 94 - 27905053 ----------------GG-------AAAUUGGCCG---UGUUGUUGGCUAUUUCCUCCUUGGCCACUUUUUAGCUUGCCGGCAUAUCAAAUUAUAUGCACACACACACACACCGCAAGCU ----------------((-------((((.(((((---......)))))))))))................(((((((..((((((......))))))...............))))))) ( -25.23) >DroEre_CAF1 9804 97 - 1 ---------GGAAAUUGGAAAUUGGAAAUUGGCUG---UAUU---GAGGAUUUCUUUCCUGGCCACUUUUUAGCUUGCCGGCAUAUCAAAUUAUAUGCAC--------ACACCGCAAGCU ---------.............((((((.(((((.---....---.((((......))))))))).))))))((((((..((((((......))))))..--------.....)))))). ( -25.50) >DroYak_CAF1 9752 96 - 1 UAUUGGCCAG---UUUGG-------AAAUUGGCUG---UGUU---GGGGAUUUCUUUCUUGGCCACUUUUUAGCUUGCCGGCAUAUCAAAUUAUAUGCAC--------ACACCGCAAGCU ...(((((((---...((-------(((((..(..---....---)..)))))))...)))))))......(((((((..((((((......))))))..--------.....))))))) ( -30.00) >DroAna_CAF1 9609 78 - 1 ------------------------------GGCUGGUGGCUU---ACGGAUUUCUUUCUUGGCCA-UUUUUAGCUCACUGGCAUAUCAAAUUAUAUGCAC--------ACACCGCAAGCU ------------------------------(((((((((((.---..(((......))).)))))-))....((....((((((((......)))))).)--------)....)).)))) ( -19.90) >consensus ________________GG_______AAAUUGGCUG___UGUU___GGGGAUUUCUUUCUUGGCCACUUUUUAGCUUGCCGGCAUAUCAAAUUAUAUGCAC________ACACCGCAAGCU ..............................((((..........................)))).......(((((((..((((((......))))))...............))))))) (-14.49 = -14.80 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:29 2006