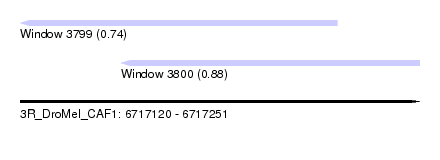
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,717,120 – 6,717,251 |
| Length | 131 |
| Max. P | 0.883140 |

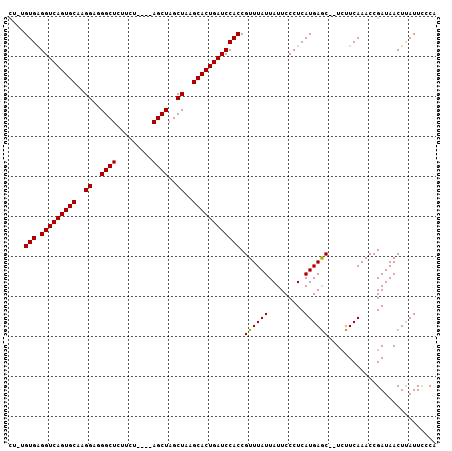
| Location | 6,717,120 – 6,717,224 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -23.72 |
| Energy contribution | -23.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6717120 104 - 27905053 U--GGUGGGGUCAGUGCAUGG-GGGCUUUGCUAGCUAGCUAGCUAAGCACUGAUCCACCGUUUAUUAUUCCCUCAUGAACACUAUUCAAACCGAUAACUCAUUUCCA .--((((((.(((((((.(((-.((((..(....).))))..))).)))))))))))))((((((.........))))))........................... ( -31.00) >DroSec_CAF1 1485 101 - 1 CUUUGUGAGGUCAGUGCAAGGAGGGCUCUUUU----AGCUAGCUAAGCACUGAUCCACCGUUUAUUAUUCCCUCAUGAGC--UCUUCAAACCGAUAACUUAUUCCCA ....(((.(((((((((..((..((((.....----))))..))..)))))))))))).((((((.........))))))--......................... ( -25.90) >DroSim_CAF1 1484 101 - 1 CUGUGUGAGGUCAGUGCAAGGAGGGCUCUUGU----AGCUAGCUAAGCACUGAUCCACCGUUUAUUAUUCCCUCAUGAGC--UCUUCAAACCGAUAACUUAUUCCCA ..(.(((.(((((((((..((..((((.....----))))..))..)))))))))))))((((((.........))))))--......................... ( -26.30) >consensus CU_UGUGAGGUCAGUGCAAGGAGGGCUCUUCU____AGCUAGCUAAGCACUGAUCCACCGUUUAUUAUUCCCUCAUGAGC__UCUUCAAACCGAUAACUUAUUCCCA ....(((.(((((((((..((..((((.........))))..))..)))))))))))).((((((.........))))))........................... (-23.72 = -23.50 + -0.22)

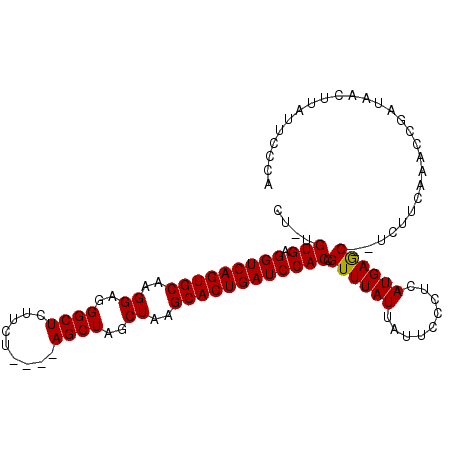
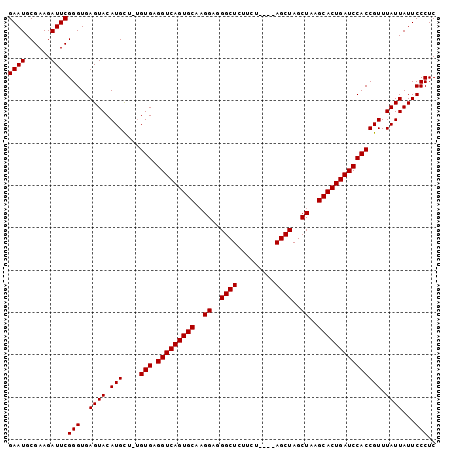
| Location | 6,717,153 – 6,717,251 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 89.97 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -30.73 |
| Energy contribution | -30.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6717153 98 - 27905053 GAAUGCGAAGAUUCGGGUGAGUACAUGU--GGUGGGGUCAGUGCAUGG-GGGCUUUGCUAGCUAGCUAGCUAAGCACUGAUCCACCGUUUAUUAUUCCCUC ((((......))))(((.(((((....(--((((((.(((((((.(((-.((((..(....).))))..))).)))))))))))))).....)))))))). ( -36.10) >DroSec_CAF1 1516 97 - 1 GAAUGCGAAGAUUCGGGUGAGUACAUGCUUUGUGAGGUCAGUGCAAGGAGGGCUCUUUU----AGCUAGCUAAGCACUGAUCCACCGUUUAUUAUUCCCUC ((((......))))(((..((((.(((....(((.(((((((((..((..((((.....----))))..))..))))))))))))))).))))...))).. ( -32.50) >DroSim_CAF1 1515 97 - 1 GAAUGCGAAGAUUCGGGUGAGUACAUGCUGUGUGAGGUCAGUGCAAGGAGGGCUCUUGU----AGCUAGCUAAGCACUGAUCCACCGUUUAUUAUUCCCUC ((((......))))(((..((((.(((....(((.(((((((((..((..((((.....----))))..))..))))))))))))))).))))...))).. ( -32.50) >consensus GAAUGCGAAGAUUCGGGUGAGUACAUGCU_UGUGAGGUCAGUGCAAGGAGGGCUCUUCU____AGCUAGCUAAGCACUGAUCCACCGUUUAUUAUUCCCUC ((((......))))(((..((((.(((....(((.(((((((((..((..((((.........))))..))..))))))))))))))).))))...))).. (-30.73 = -30.73 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:16 2006