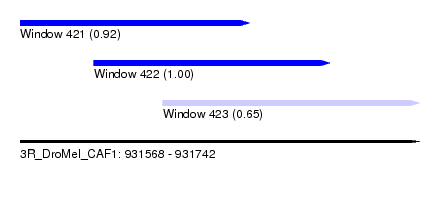
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 931,568 – 931,742 |
| Length | 174 |
| Max. P | 0.998007 |

| Location | 931,568 – 931,668 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 71.72 |
| Mean single sequence MFE | -21.54 |
| Consensus MFE | -11.71 |
| Energy contribution | -11.02 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 931568 100 + 27905053 AUAAUAUCAGGCUUUCAACGAUAAAAAAAGGGGGGCUACUUUAAUUAGUUGUAGCAGCUUUA------------AUGAACAACGAAAAAUGCAGGAAACUGCAAAAUGUUUA ......((.((((..(..............)..))))..........(((((..((......------------.)).)))))))....(((((....)))))......... ( -20.84) >DroSec_CAF1 20597 99 + 1 GUAAUAUCAAGCUUUCAACGAUGAA-AAAGAGGGGCUACUUUAAUUAAUUGUAGCAGCCAUA------------AUGAACAGCGAAAAACGCAGGAAACUGCACAAUGUUUA ..........(((((((....(...-.)...((.(((((...........)))))..))...------------.)))).))).......((((....)))).......... ( -22.00) >DroSim_CAF1 26510 97 + 1 --AAUAUCAAGCUUUCAACAAUGAA-AAAGAGGGGCUGCUUUAAUUAAUUGUAGCAGUUAUA------------AUGAACAACGAAAAACGCAGGAAACUGCAAAAUGUUUA --....(((...(((((....))))-)......(((((((.(((....))).)))))))...------------.)))............((((....)))).......... ( -20.10) >DroEre_CAF1 21614 100 + 1 GUAAUGCCAGGUUUGCAGCGGUAA--AAAGGGGAAUUACCUAAAUUAAUUGUAAUAGCUAUAGCGUUGUUGCUGAACAGCG----------CAGGAAAGUGCAGCAUUUUUA ..(((((...(((((((((((...--...(((......)))...............((....)).))))))).)))).(((----------(......)))).))))).... ( -23.20) >consensus GUAAUAUCAAGCUUUCAACGAUAAA_AAAGAGGGGCUACUUUAAUUAAUUGUAGCAGCUAUA____________AUGAACAACGAAAAACGCAGGAAACUGCAAAAUGUUUA .........((((....(((((......((((......)))).....)))))...))))...............................((((....)))).......... (-11.71 = -11.02 + -0.69)

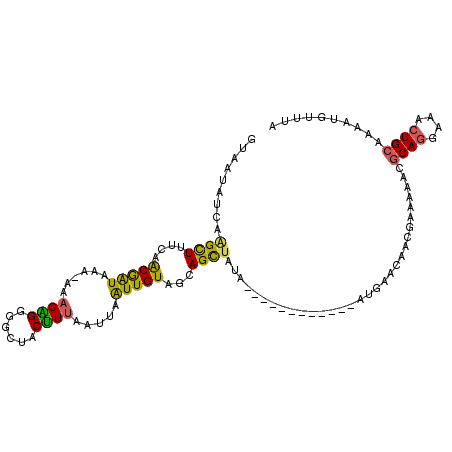
| Location | 931,600 – 931,703 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.35 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -14.26 |
| Energy contribution | -14.20 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 931600 103 + 27905053 GGGCUACUUUAAUUAGUUGUAGCAGCUUUA------------AUGAACAACGAAAAAUGCAGGAAACUGCAAAAUGUUUACUACUCUUAAGUCUAAACUCAGACUCCAG---CU-UUUU ..(((((...........)))))((((...------------.((((((........(((((....)))))...)))))).........(((((......)))))..))---))-.... ( -23.30) >DroSec_CAF1 20628 107 + 1 GGGCUACUUUAAUUAAUUGUAGCAGCCAUA------------AUGAACAGCGAAAAACGCAGGAAACUGCACAAUGUUUACAAGUCUUUAGUCUAAACACAGACUACAAAAACUGGCUA ..(((((...........)))))(((((..------------.((((((..(.....)((((....))))....))))))........((((((......)))))).......))))). ( -29.00) >DroSim_CAF1 26539 107 + 1 GGGCUGCUUUAAUUAAUUGUAGCAGUUAUA------------AUGAACAACGAAAAACGCAGGAAACUGCAAAAUGUUUACUAGUCUUUAGUCUAAACGCAGACUACAAAAACUGGUUA .(((((((.(((....))).)))))))...------------.((((((..(.....)((((....))))....))))))(((((.((((((((......))))))..)).)))))... ( -27.80) >DroEre_CAF1 21644 106 + 1 GAAUUACCUAAAUUAAUUGUAAUAGCUAUAGCGUUGUUGCUGAACAGCG----------CAGGAAAGUGCAGCAUUUUUAUUACUCAUUAAUCUAUACUUAGACUGCAGAACCUGG--- ......((......................((((((((....)))))))----------)(((....(((((.....(((........)))((((....)))))))))...)))))--- ( -19.60) >consensus GGGCUACUUUAAUUAAUUGUAGCAGCUAUA____________AUGAACAACGAAAAACGCAGGAAACUGCAAAAUGUUUACUACUCUUUAGUCUAAACUCAGACUACAAAAACUGGUUA ..(((((...........)))))...................................((((....))))..................((((((......))))))............. (-14.26 = -14.20 + -0.06)


| Location | 931,630 – 931,742 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.07 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -16.51 |
| Energy contribution | -16.40 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 931630 112 + 27905053 AUGAACAACGAAAAAUGCAGGAAACUGCAAAAUGUUUACUACUCUUAAGUCUAAACUCAGACUCCAG---CU-UUUUCAUCAGUGGCUUAUCCAUUUCAAAGAAAUAAUAAGGUUU ..((((...(((((((((((....)))))..............((..(((((......)))))..))---.)-)))))........(((((..(((((...))))).))))))))) ( -22.20) >DroSec_CAF1 20658 116 + 1 AUGAACAGCGAAAAACGCAGGAAACUGCACAAUGUUUACAAGUCUUUAGUCUAAACACAGACUACAAAAACUGGCUACAUGCAUGGCUUAUUCAUUUUAAAGAAAUAAUAAGGUUU ..((((.((.......((((....))))...((((.....(((.((((((((......))))))..)).)))....))))))....((((((.(((((...))))))))))))))) ( -24.40) >DroSim_CAF1 26569 116 + 1 AUGAACAACGAAAAACGCAGGAAACUGCAAAAUGUUUACUAGUCUUUAGUCUAAACGCAGACUACAAAAACUGGUUACAUGCAUGGCAUAUUCAUUUUAAAGAAAUAAUAAGGUUU (((((...........((((....))))...((((..((((((.((((((((......))))))..)).)))))).))))..........)))))..................... ( -24.40) >consensus AUGAACAACGAAAAACGCAGGAAACUGCAAAAUGUUUACUAGUCUUUAGUCUAAACACAGACUACAAAAACUGGUUACAUGCAUGGCUUAUUCAUUUUAAAGAAAUAAUAAGGUUU ..((((..........((((....))))..............((((((((((......)))))...................((((.....))))...))))).........)))) (-16.51 = -16.40 + -0.11)

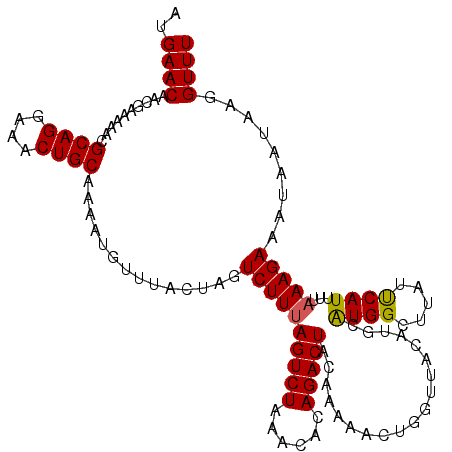
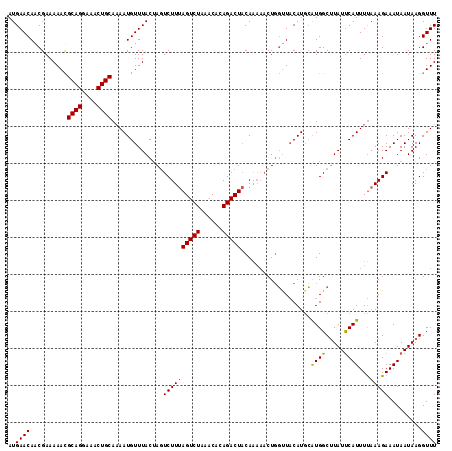
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:25 2006