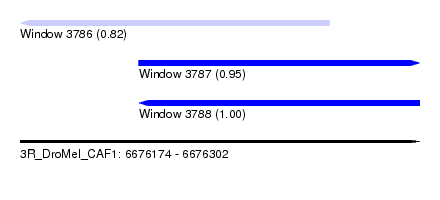
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,676,174 – 6,676,302 |
| Length | 128 |
| Max. P | 0.995621 |

| Location | 6,676,174 – 6,676,273 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -19.41 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6676174 99 - 27905053 AUAC-----------GUAUGCGAUGAUGAUAUACCAUCGAAAGUGGAUACUGCUACUUUCAGAUAGA--------AUGUGUCUGGGUACGGAUUCUCGUGCGGUUUGUUUACCUGCUC-- ...(-----------((((.(...((((......))))((((((((......))))))))(((((..--------...)))))).))))).........(((((......))).))..-- ( -25.80) >DroSec_CAF1 60776 99 - 1 GUAC-----------GGAUGCGAUGAUGAUAUACCAUCGAAAGUGGAUACUGCUACUUUCAGGUAGA--------AUGUGUCUGGGUACGGAUUCUCGUGCGGUUUGUUUACCUGCUC-- ...(-----------(((((((..((((......))))((((((((......)))))))).......--------.)))))))).((((((....))))))(((......))).....-- ( -30.20) >DroSim_CAF1 61772 99 - 1 GUAC-----------GGAUGCGAUGAUGAUAUACCAUCGAAAGUGGAUACUGCUACUUUCAGGUAGA--------AUGUGUCUGGGUACGGAUUCUCGUGCGGUUUGUUUACCUGCUC-- ...(-----------(((((((..((((......))))((((((((......)))))))).......--------.)))))))).((((((....))))))(((......))).....-- ( -30.20) >DroEre_CAF1 61940 109 - 1 GUAU-----------GGAUGCGAUGAUGAUACACCAUCGAAAGUGGAUACUGAUACUUUCAGGUAGAAUAACCGAAUGGAUCUGAGUACAGAUUCUCGUGCGGUUUGUUUACCUGCUCUC ....-----------(((.((...((((......))))(((((((........)))))))(((((((..(((((...(((((((....))))))).....)))))..)))))))))))). ( -35.00) >DroYak_CAF1 62469 107 - 1 GUAU-----------GGAUGCGAUGAUGAUAUACCAUCGAAAGUGGAUACUGAUACUUCCAGGUAGAAGGGCUGAAUGGAUCUGAGUACAGAUUCUCGUGCGGUUUGUUUACCUGCUC-- ((((-----------..((.(((((.........)))))...))..)))).........((((((((..(((((...(((((((....))))))).....)))))..))))))))...-- ( -31.70) >DroAna_CAF1 67175 103 - 1 GAAAAAAAAGAAGUAAAAAGCGAUGAGAAUACAACGCCGAAAGUGGAAAC----ACUUUCAGGUAGA--------ACCAAUCUGAGAGCGGGUUU---CGAGGUUUGUUUACCUGCAC-- ...................(((.((......)).))).(((((((....)----))))))(((((((--------...(((((((((.....)))---).)))))..)))))))....-- ( -27.70) >consensus GUAC___________GGAUGCGAUGAUGAUAUACCAUCGAAAGUGGAUACUGCUACUUUCAGGUAGA________AUGUAUCUGAGUACGGAUUCUCGUGCGGUUUGUUUACCUGCUC__ ...................((((((.........))))(((((((........)))))))(((((((........(((.(((((....)))))...)))........))))))))).... (-19.41 = -19.82 + 0.42)

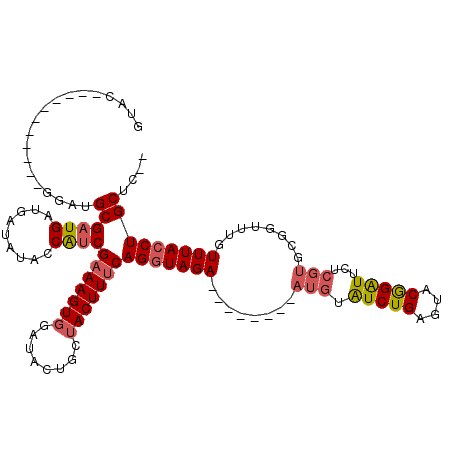
| Location | 6,676,212 – 6,676,302 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.32 |
| Mean single sequence MFE | -20.09 |
| Consensus MFE | -10.66 |
| Energy contribution | -10.92 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6676212 90 + 27905053 CACAU--------UCUAUCUGAAAGUAGCAGUAUCCACUUUCGAUGGUAUAUCAUCAUCGCAUAC-----------GUAUA------AACAUUC-----AGCUGGUAGAAUGAAUUACUU ..(((--------((((((.((((((.(......).))))))((((((.....))))))......-----------.....------.......-----....)))))))))........ ( -20.40) >DroSec_CAF1 60814 90 + 1 CACAU--------UCUACCUGAAAGUAGCAGUAUCCACUUUCGAUGGUAUAUCAUCAUCGCAUCC-----------GUACA------AACAUUC-----AGCUGGCAGAAUGAAUUACUU ..(((--------(((.((.((((((.(......).))))))((((((.....))))))......-----------.....------.......-----....)).))))))........ ( -18.40) >DroSim_CAF1 61810 90 + 1 CACAU--------UCUACCUGAAAGUAGCAGUAUCCACUUUCGAUGGUAUAUCAUCAUCGCAUCC-----------GUACA------AACAUUU-----AGCUGGCAGAAUGAAUUACUU ..(((--------(((.((.((((((.(......).))))))((((((.....))))))......-----------.....------.......-----....)).))))))........ ( -18.40) >DroEre_CAF1 61980 102 + 1 UCCAUUCGGUUAUUCUACCUGAAAGUAUCAGUAUCCACUUUCGAUGGUGUAUCAUCAUCGCAUCC-----------AUACAAA--CAAACAUUC-----AGAUGGCAGAAUGAAUUACUU ..(((((.((((((......((((((..........))))))(((((((...)))))))......-----------.......--.........-----.)))))).)))))........ ( -20.10) >DroYak_CAF1 62507 104 + 1 UCCAUUCAGCCCUUCUACCUGGAAGUAUCAGUAUCCACUUUCGAUGGUAUAUCAUCAUCGCAUCC-----------AUACAAACACAAACAUUC-----AGCUGGCAGAAUGAAUUACUU ..(((((.((((((((....))))).....((((.......(((((((.....))))))).....-----------))))..............-----....))).)))))........ ( -20.70) >DroAna_CAF1 67210 106 + 1 UUGGU--------UCUACCUGAAAGU----GUUUCCACUUUCGGCGUUGUAUUCUCAUCGCUUUUUACUUCUUUUUUUUCAAG--CUAAUGCUCAUACAAGCUGGUCGAAUAAAUUACUU ...((--------((.(((.((((((----(....)))))))(((.((((((...(((.((((.................)))--)..)))...)))))))))))).))))......... ( -22.53) >consensus CACAU________UCUACCUGAAAGUAGCAGUAUCCACUUUCGAUGGUAUAUCAUCAUCGCAUCC___________GUACA______AACAUUC_____AGCUGGCAGAAUGAAUUACUU .............(((.((.((((((..........))))))((((((.....))))))............................................)).)))........... (-10.66 = -10.92 + 0.25)

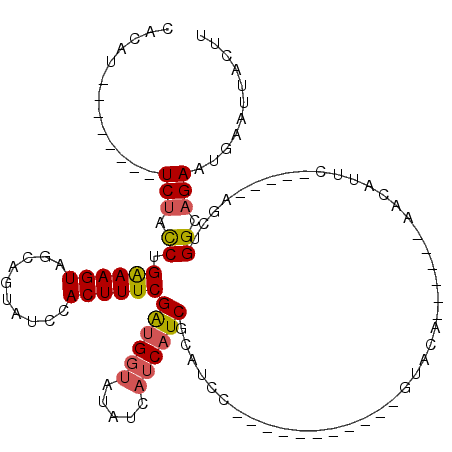
| Location | 6,676,212 – 6,676,302 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.32 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -14.58 |
| Energy contribution | -15.05 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6676212 90 - 27905053 AAGUAAUUCAUUCUACCAGCU-----GAAUGUU------UAUAC-----------GUAUGCGAUGAUGAUAUACCAUCGAAAGUGGAUACUGCUACUUUCAGAUAGA--------AUGUG ........(((((((.(.((.-----..((((.------...))-----------))..))...((((......))))((((((((......)))))))).).))))--------))).. ( -22.50) >DroSec_CAF1 60814 90 - 1 AAGUAAUUCAUUCUGCCAGCU-----GAAUGUU------UGUAC-----------GGAUGCGAUGAUGAUAUACCAUCGAAAGUGGAUACUGCUACUUUCAGGUAGA--------AUGUG ........(((((((((.((.-----...(((.------...))-----------)...))...((((......))))((((((((......)))))))).))))))--------))).. ( -28.40) >DroSim_CAF1 61810 90 - 1 AAGUAAUUCAUUCUGCCAGCU-----AAAUGUU------UGUAC-----------GGAUGCGAUGAUGAUAUACCAUCGAAAGUGGAUACUGCUACUUUCAGGUAGA--------AUGUG ........(((((((((.((.-----...(((.------...))-----------)...))...((((......))))((((((((......)))))))).))))))--------))).. ( -28.40) >DroEre_CAF1 61980 102 - 1 AAGUAAUUCAUUCUGCCAUCU-----GAAUGUUUG--UUUGUAU-----------GGAUGCGAUGAUGAUACACCAUCGAAAGUGGAUACUGAUACUUUCAGGUAGAAUAACCGAAUGGA ..(((..(((((...((((..-----((((....)--)))..))-----------))....)))))...))).((((((((((((........))))))).(((......)))).)))). ( -23.70) >DroYak_CAF1 62507 104 - 1 AAGUAAUUCAUUCUGCCAGCU-----GAAUGUUUGUGUUUGUAU-----------GGAUGCGAUGAUGAUAUACCAUCGAAAGUGGAUACUGAUACUUCCAGGUAGAAGGGCUGAAUGGA ......(((((((.(((..((-----(..((...(((((.((((-----------..((.(((((.........)))))...))..)))).)))))...))..)))...))).))))))) ( -30.10) >DroAna_CAF1 67210 106 - 1 AAGUAAUUUAUUCGACCAGCUUGUAUGAGCAUUAG--CUUGAAAAAAAAGAAGUAAAAAGCGAUGAGAAUACAACGCCGAAAGUGGAAAC----ACUUUCAGGUAGA--------ACCAA ..........(((.(((.((((((((...((((.(--(((.................))))))))...)))))).)).(((((((....)----)))))).))).))--------).... ( -30.33) >consensus AAGUAAUUCAUUCUGCCAGCU_____GAAUGUU______UGUAC___________GGAUGCGAUGAUGAUAUACCAUCGAAAGUGGAUACUGCUACUUUCAGGUAGA________AUGUA ...........((((((............................................((((.........))))(((((((........))))))).))))))............. (-14.58 = -15.05 + 0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:05 2006