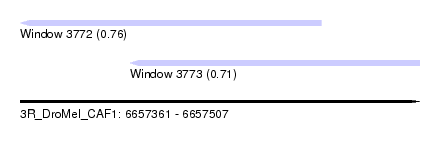
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,657,361 – 6,657,507 |
| Length | 146 |
| Max. P | 0.756103 |

| Location | 6,657,361 – 6,657,471 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.23 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6657361 110 - 27905053 UGCUUUCUAAGAAUUUUCAUUGUACUUGCGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAAGUAAUGUGUAUGCUUUAAAGUGUCUCGAAGGCUUGCCUUUCAUUUA .(((((((((((((((.((.......)).))))))))((......))...)))))))((((((..(((.((....)).)))...))))))(((((....)))))...... ( -30.30) >DroSim_CAF1 42575 109 - 1 UGCUUUCUAAGAAUUUUCAUUGUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUGUGUGUGCUUUAAAGUGUCUCGAAGGCAAGCCUUUCAUUUA .(((((((((((((((.((.......)).))))))))((......))...)))))))((((((.-(((.((....)).)))...))))))(((((....)))))...... ( -28.80) >DroEre_CAF1 43153 109 - 1 UGCUUUCUAAGAAUUUUCUUUGUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUGUGUGUGCUUUAAAGUGUCUCGAAGUCUCGCCUUUUAUUUA ...(((((((((((((.(.........).)))))))))).)))....((..(((((((((((..-...((.(.(..((....))..)).))..)))))))))))..)).. ( -26.40) >DroYak_CAF1 43177 109 - 1 UGCUUUCUAAGAAUUUUCAUUGUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUGUGUGUGCUUCAAAGUGUCUCGAAGUCUCGCCUUUUAUUUA ...(((((((((((((.((.......)).)))))))))).)))....((..(((((((((((..-...((.(.(..((....))..)).))..)))))))))))..)).. ( -28.10) >consensus UGCUUUCUAAGAAUUUUCAUUGUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA_UAAUGUGUGUGCUUUAAAGUGUCUCGAAGGCUCGCCUUUCAUUUA ...(((((((((((((.((.......)).)))))))))).))).......(((((((((((((..(((.((....)).)))...))))))(....)..)))))))..... (-25.98 = -26.23 + 0.25)


| Location | 6,657,401 – 6,657,507 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 96.52 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -23.36 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6657401 106 - 27905053 CCUAUCAGCAAAUCAAUGUUGACUCAAAUACCUUGGUGCUUUCUAAGAAUUUUCAUUGUACUUGCGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAAGUAAUG (((.((((((......))))))........((((((((.((.((((((((((.((.......)).)))))))))).)).)))))).)))))............... ( -27.30) >DroSim_CAF1 42615 105 - 1 CCUAUCAGCAAAUCAAUGUUGACUCAAAUACCUUGGUGCUUUCUAAGAAUUUUCAUUGUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUG (((.((((((......))))))........((((((((.((.((((((((((.((.......)).)))))))))).)).)))))).))))).........-..... ( -27.30) >DroEre_CAF1 43193 105 - 1 CCUAUCUGCAAAUCAAUGUUGACUCAAAUACCGUGGUGCUUUCUAAGAAUUUUCUUUGUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUG ................(((((.(((.....((((((((.((.((((((((((.(.........).)))))))))).)).)))).)))).....))).)))-))... ( -25.20) >DroYak_CAF1 43217 105 - 1 CCUAUCUGCAAAUCAAUGUCGGCUCAAAUACCUUGGUGCUUUCUAAGAAUUUUCAUUGUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUG ................((((.((((.....((((((((.((.((((((((((.((.......)).)))))))))).)).)))))).)).))))..)))).-..... ( -27.60) >consensus CCUAUCAGCAAAUCAAUGUUGACUCAAAUACCUUGGUGCUUUCUAAGAAUUUUCAUUGUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA_UAAUG (((.((((((......))))))........((((((((.((.((((((((((.((.......)).)))))))))).)).)))))).)))))............... (-23.36 = -24.18 + 0.81)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:50 2006