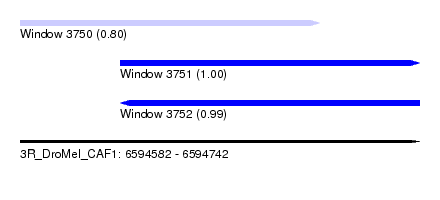
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,594,582 – 6,594,742 |
| Length | 160 |
| Max. P | 0.995112 |

| Location | 6,594,582 – 6,594,702 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.84 |
| Mean single sequence MFE | -47.37 |
| Consensus MFE | -41.71 |
| Energy contribution | -41.93 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6594582 120 + 27905053 CAUCUUCAGCGGCUUUUAGGGCUUUAGCUUCUUCGGCUGCUUUUUGGGCGGCAGCUUCAGCCAGGAGAGCCUCUUCGGCAGCGAUUUUGGCGGCCUCCUCGGCAGCCUUUUUGGCCUUAG ..........((((...((((((..((((......((((((.....))))))))))...((((((((.(((.(((((....)))....)).)))))))).)))))))))...)))).... ( -46.10) >DroSec_CAF1 7038 117 + 1 CAUCUUCAGCGGCUUUUAAGGC---AGCUUCUUCGGCAGCUUUUUGGGCUGCAGCUUCGGCCAGGAGAGCCUCUUCGGCUGCGAUUUUGGCGGCCUCCUCGGCAGCCUUUUUGGCCUUGG ......(((.((((...(((((---....(((((.((((((.....)))))).((....))..)))))(((.....((((((.......)))))).....))).)))))...))))))). ( -50.10) >DroSim_CAF1 7858 117 + 1 CAUCUUCAGCUGCUUUUAAGGC---AGCUUCUUCUGCAGCUUUUUGGGCGGCAGCUUCGGCCAGGAGAGCCUCUUCGGCAGCGAUUUUGGCGGCCUCCUCGGCAGCCUUUUUGGCCUUGG ........(((((((..(((((---.((.......)).)))))..)))))))......(((((((((.(((.....))).........(((.(((.....))).)))))))))))).... ( -45.90) >consensus CAUCUUCAGCGGCUUUUAAGGC___AGCUUCUUCGGCAGCUUUUUGGGCGGCAGCUUCGGCCAGGAGAGCCUCUUCGGCAGCGAUUUUGGCGGCCUCCUCGGCAGCCUUUUUGGCCUUGG ..........((((...(((((...((((......((((((.....))))))))))...((((((((.(((.(((((....)))....)).)))))))).))).)))))...)))).... (-41.71 = -41.93 + 0.23)


| Location | 6,594,622 – 6,594,742 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -52.08 |
| Consensus MFE | -43.68 |
| Energy contribution | -44.36 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6594622 120 + 27905053 UUUUUGGGCGGCAGCUUCAGCCAGGAGAGCCUCUUCGGCAGCGAUUUUGGCGGCCUCCUCGGCAGCCUUUUUGGCCUUAGCAGCAGCCUGCUUUUCGGCCUCAGCCGUCAGGCGAUCCCU .....(((.(((.(((.(.((((((((.(((.(((((....)))....)).)))))))).))).(((.....)))....).))).)))(((((..((((....))))..)))))..))). ( -49.00) >DroSec_CAF1 7075 120 + 1 UUUUUGGGCUGCAGCUUCGGCCAGGAGAGCCUCUUCGGCUGCGAUUUUGGCGGCCUCCUCGGCAGCCUUUUUGGCCUUGGCAGCAGCCAGCUUUUCGGCCUCAGCCGUCAGGCGAUCCCU ......((((((.(((..(((((((((((((.....))))........(((.(((.....))).))))))))))))..))).)))))).((((..((((....))))..))))....... ( -56.70) >DroSim_CAF1 7895 120 + 1 UUUUUGGGCGGCAGCUUCGGCCAGGAGAGCCUCUUCGGCAGCGAUUUUGGCGGCCUCCUCGGCAGCCUUUUUGGCCUUGGCAGCAGCCUGCUUUUCGGCCUCAGCCGUCAGGCGAUCCCU .....((((.((.(((..(((((((((.(((.....))).........(((.(((.....))).))))))))))))..))).)).))))((((..((((....))))..))))....... ( -51.90) >DroEre_CAF1 7450 120 + 1 UUUUUGGGCGGCAGCUUCGGCCAGGAGAGCCUCUUCGGCAGCGAUCUUGGCGGCUUCCUCGGCCGCCUUUUUGGCCUUGGCAGCAGCCUGCUUCUCGGCCUCAGCUGUCAGACGUUCCCU ....(((.(((.....))).)))((.((((.(((..((((((......(((((((.....))))))).....((((..(((((....)))))....))))...))))))))).)))))). ( -51.80) >DroYak_CAF1 7475 120 + 1 UUUUUGUGCGGCAGCUUCGGCCUGGACAGCUGCCUCGGCAGCAAUUUUGGCGGCCUCCUCGGCAGCCUUUUUGGCCUUGGCAGCAGCCUGCUUCUCGGCCUCAGCUGUCAGACGUUCCCU .....((..(((((((..((((.(((..((((((..(((..(((....(((.(((.....))).)))...))))))..))))))((....))))).))))..)))))))..))....... ( -51.00) >consensus UUUUUGGGCGGCAGCUUCGGCCAGGAGAGCCUCUUCGGCAGCGAUUUUGGCGGCCUCCUCGGCAGCCUUUUUGGCCUUGGCAGCAGCCUGCUUUUCGGCCUCAGCCGUCAGGCGAUCCCU .....((((.((.(((..(((((((((.(((.....))).........(((.(((.....))).))))))))))))..))).)).))))((((..((((....))))..))))....... (-43.68 = -44.36 + 0.68)


| Location | 6,594,622 – 6,594,742 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -50.66 |
| Consensus MFE | -40.94 |
| Energy contribution | -41.34 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6594622 120 - 27905053 AGGGAUCGCCUGACGGCUGAGGCCGAAAAGCAGGCUGCUGCUAAGGCCAAAAAGGCUGCCGAGGAGGCCGCCAAAAUCGCUGCCGAAGAGGCUCUCCUGGCUGAAGCUGCCGCCCAAAAA .(((...(((((.((((....)))).....))))).((.(((..(((((....(((.(((.....))).))).........(((.....))).....)))))..))).))..)))..... ( -50.30) >DroSec_CAF1 7075 120 - 1 AGGGAUCGCCUGACGGCUGAGGCCGAAAAGCUGGCUGCUGCCAAGGCCAAAAAGGCUGCCGAGGAGGCCGCCAAAAUCGCAGCCGAAGAGGCUCUCCUGGCCGAAGCUGCAGCCCAAAAA .(((.....(((.(((((..(((((...((((((((((.((....))......(((.(((.....))).)))......)))))).....))))....)))))..)))))))))))..... ( -51.90) >DroSim_CAF1 7895 120 - 1 AGGGAUCGCCUGACGGCUGAGGCCGAAAAGCAGGCUGCUGCCAAGGCCAAAAAGGCUGCCGAGGAGGCCGCCAAAAUCGCUGCCGAAGAGGCUCUCCUGGCCGAAGCUGCCGCCCAAAAA .(((...(((((.((((....)))).....))))).((.((...(((((....(((.(((.....))).))).........(((.....))).....)))))...)).))..)))..... ( -50.20) >DroEre_CAF1 7450 120 - 1 AGGGAACGUCUGACAGCUGAGGCCGAGAAGCAGGCUGCUGCCAAGGCCAAAAAGGCGGCCGAGGAAGCCGCCAAGAUCGCUGCCGAAGAGGCUCUCCUGGCCGAAGCUGCCGCCCAAAAA .(((......((.(((((..(((((((((((.((((........)))).....((((((.......))))))......)))(((.....)))))))..))))..))))).)))))..... ( -50.80) >DroYak_CAF1 7475 120 - 1 AGGGAACGUCUGACAGCUGAGGCCGAGAAGCAGGCUGCUGCCAAGGCCAAAAAGGCUGCCGAGGAGGCCGCCAAAAUUGCUGCCGAGGCAGCUGUCCAGGCCGAAGCUGCCGCACAAAAA .(....)((.((.(((((..(((((.....).(((.((((((..((((((...(((.(((.....))).)))....)))..)))..)))))).)))..))))..))))).)).))..... ( -50.10) >consensus AGGGAUCGCCUGACGGCUGAGGCCGAAAAGCAGGCUGCUGCCAAGGCCAAAAAGGCUGCCGAGGAGGCCGCCAAAAUCGCUGCCGAAGAGGCUCUCCUGGCCGAAGCUGCCGCCCAAAAA .(((.......(.(((((..((((....(((.((((........)))).....(((.(((.....))).)))......)))(((.....)))......))))..))))))..)))..... (-40.94 = -41.34 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:30 2006