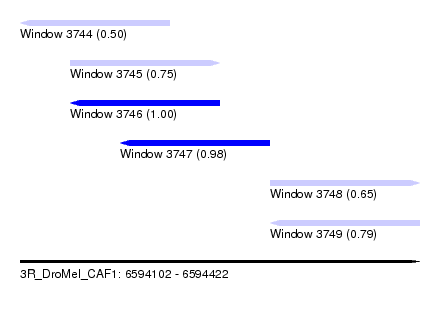
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,594,102 – 6,594,422 |
| Length | 320 |
| Max. P | 0.996822 |

| Location | 6,594,102 – 6,594,222 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -49.50 |
| Consensus MFE | -47.22 |
| Energy contribution | -46.94 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6594102 120 - 27905053 UGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCGCCAGGCCGCCGAGCUGGCCGAGAUCGCCCGCCAGGAAUCCGAGCUUGCCGCCCAGGAACUGCAGGCCAUCCAGAAGAACG ((((((.((((.....((.(((..((......))..))).)).((((((...)).))))..))))..))))))(((....(.((((((..(....)....))))))).)))......... ( -48.50) >DroSec_CAF1 6558 120 - 1 UGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCGCCAGGCCGCCGAGCUGGCCGAGAUCGCCCGCCAGGAAUCCGAGCUUGCCGCCCAGGAGCUGCAGGCUAUCCAGAAGAACG ((((((.((((.....((.(((..((......))..))).)).((((((...)).))))..))))..))))))(((.....(((((((.((......)).))))))).)))......... ( -54.00) >DroSim_CAF1 7379 120 - 1 UGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCGCCAGGCCGCCGAGCUGGCCGAGAUCGCCCGCCAGGAAUCCGAGCUUGCCGCCCAGGAGCUGCAGGCCAUCCAGAAGAACG ((((((.((((.....((.(((..((......))..))).)).((((((...)).))))..))))..))))))(((....(.((((((.((......)).))))))).)))......... ( -51.70) >DroEre_CAF1 6933 120 - 1 UGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCGCCAGGCCGCCGAGCUGGCCGAGAUUGCCCGCCAGGAAUCCGAGCUUGCCGCCCAGGAACUGCAGGCCAUCCAAAAGAACG ((((((.((((.....((.(((..((......))..))).)).((((((...)).))))..))))..))))))(((....(.((((((..(....)....))))))).)))......... ( -46.60) >DroYak_CAF1 6958 120 - 1 UGGCGGAGAUCGAGAAGGAGAGCGAGGGAGAACUUGCUCGCCAAGCCGCCGAGCUGGCCGAGAUCGCCCGCCAGGAAUCCGAGCUUGCCGCCCAGGAACUGCAGGCCAUCCAGAAGAACG ((((((.((((..(..((.(((((((......))))))).))..(((((...)).))))..))))..))))))(((....(.((((((..(....)....))))))).)))......... ( -46.70) >consensus UGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCGCCAGGCCGCCGAGCUGGCCGAGAUCGCCCGCCAGGAAUCCGAGCUUGCCGCCCAGGAACUGCAGGCCAUCCAGAAGAACG ((((((.((((.....((.(((((((......))))))).)).((((((...)).))))..))))..))))))(((....(.((((((.(........).))))))).)))......... (-47.22 = -46.94 + -0.28)


| Location | 6,594,142 – 6,594,262 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -44.08 |
| Consensus MFE | -41.44 |
| Energy contribution | -41.68 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6594142 120 + 27905053 GGAUUCCUGGCGGGCGAUCUCGGCCAGCUCGGCGGCCUGGCGAGCAAGUUCUCCCUCACUCUCCUUCUCGAUCUCCGCCAGGCGCUCCAGCUCCUGCUCACGGACACGCUGCUCCUCCUC (((..(((((((((.((((..((((.((...)))))).((.(((..((......))..))).)).....))))))))))))).....(((((((.......)))...)))).)))..... ( -44.40) >DroSec_CAF1 6598 120 + 1 GGAUUCCUGGCGGGCGAUCUCGGCCAGCUCGGCGGCCUGGCGAGCAAGUUCUCCCUCACUCUCCUUCUCGAUCUCCGCCAGGCGCUCCAGCUCCUGCUCACGGACACGCUGCUCCUCCUC (((..(((((((((.((((..((((.((...)))))).((.(((..((......))..))).)).....))))))))))))).....(((((((.......)))...)))).)))..... ( -44.40) >DroSim_CAF1 7419 120 + 1 GGAUUCCUGGCGGGCGAUCUCGGCCAGCUCGGCGGCCUGGCGAGCAAGUUCUCCCUCACUCUCCUUCUCGAUCUCCGCCAGGCGCUCCAGCUCCUGCUCACGGACACGCUGCUCCUCCUC (((..(((((((((.((((..((((.((...)))))).((.(((..((......))..))).)).....))))))))))))).....(((((((.......)))...)))).)))..... ( -44.40) >DroEre_CAF1 6973 120 + 1 GGAUUCCUGGCGGGCAAUCUCGGCCAGCUCGGCGGCCUGGCGAGCAAGUUCUCCCUCACUCUCCUUCUCGAUCUCCGCCAGGCGCUCCAGCUCCUGCUCACGCACCCGCUGCUCCUCCUC (((..(((((((((..(((..((((.((...)))))).((.(((..((......))..))).)).....))).))))))))).....((((...(((....)))...)))).)))..... ( -41.10) >DroYak_CAF1 6998 120 + 1 GGAUUCCUGGCGGGCGAUCUCGGCCAGCUCGGCGGCUUGGCGAGCAAGUUCUCCCUCGCUCUCCUUCUCGAUCUCCGCCAGGCGCUCCAGCUCCUGCUCACGGACACGCUGCUCCUCCUC (((..(((((((((.((((...(((((((....))).))))((((.((......)).))))........))))))))))))).....(((((((.......)))...)))).)))..... ( -46.10) >consensus GGAUUCCUGGCGGGCGAUCUCGGCCAGCUCGGCGGCCUGGCGAGCAAGUUCUCCCUCACUCUCCUUCUCGAUCUCCGCCAGGCGCUCCAGCUCCUGCUCACGGACACGCUGCUCCUCCUC (((..(((((((((.((((..((((.((...)))))).((.(((..((......))..))).)).....))))))))))))).....(((((((.......)))...)))).)))..... (-41.44 = -41.68 + 0.24)

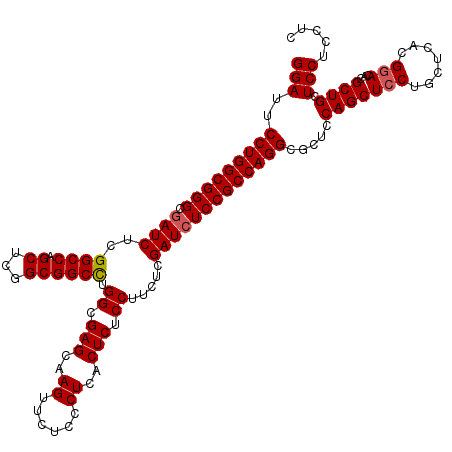

| Location | 6,594,142 – 6,594,262 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -54.40 |
| Consensus MFE | -52.72 |
| Energy contribution | -52.60 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6594142 120 - 27905053 GAGGAGGAGCAGCGUGUCCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCGCCAGGCCGCCGAGCUGGCCGAGAUCGCCCGCCAGGAAUCC .....(((.((((...(((.......))))))).....((((((((.((((.....((.(((..((......))..))).)).((((((...)).))))..))))..))))))))..))) ( -54.10) >DroSec_CAF1 6598 120 - 1 GAGGAGGAGCAGCGUGUCCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCGCCAGGCCGCCGAGCUGGCCGAGAUCGCCCGCCAGGAAUCC .....(((.((((...(((.......))))))).....((((((((.((((.....((.(((..((......))..))).)).((((((...)).))))..))))..))))))))..))) ( -54.10) >DroSim_CAF1 7419 120 - 1 GAGGAGGAGCAGCGUGUCCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCGCCAGGCCGCCGAGCUGGCCGAGAUCGCCCGCCAGGAAUCC .....(((.((((...(((.......))))))).....((((((((.((((.....((.(((..((......))..))).)).((((((...)).))))..))))..))))))))..))) ( -54.10) >DroEre_CAF1 6973 120 - 1 GAGGAGGAGCAGCGGGUGCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCGCCAGGCCGCCGAGCUGGCCGAGAUUGCCCGCCAGGAAUCC .....(((.((((...(((....)))...)))).....((((((((.((((.....((.(((..((......))..))).)).((((((...)).))))..))))..))))))))..))) ( -57.40) >DroYak_CAF1 6998 120 - 1 GAGGAGGAGCAGCGUGUCCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGCGAGGGAGAACUUGCUCGCCAAGCCGCCGAGCUGGCCGAGAUCGCCCGCCAGGAAUCC .....(((.((((...(((.......))))))).....((((((((.((((..(..((.(((((((......))))))).))..(((((...)).))))..))))..))))))))..))) ( -52.30) >consensus GAGGAGGAGCAGCGUGUCCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCGCCAGGCCGCCGAGCUGGCCGAGAUCGCCCGCCAGGAAUCC .....(((.((((.....(....).....)))).....((((((((.((((.....((.(((((((......))))))).)).((((((...)).))))..))))..))))))))..))) (-52.72 = -52.60 + -0.12)

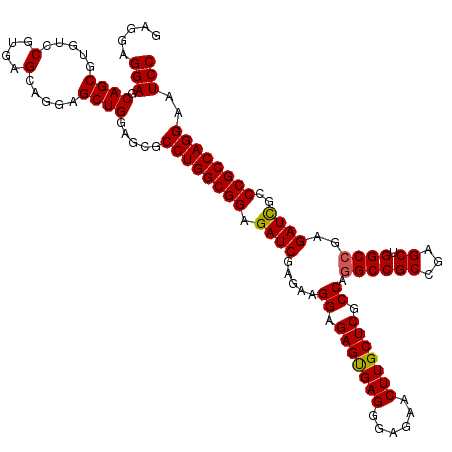

| Location | 6,594,182 – 6,594,302 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -47.34 |
| Consensus MFE | -45.58 |
| Energy contribution | -45.50 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6594182 120 - 27905053 GGCCGCCGAAGAGGCUCGUGUCGCUGAAGAGGCUCGCCUGGAGGAGGAGCAGCGUGUCCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCG ..((((((....(((((.((((((.((.(..((((.((....))..))))..)...)).)))).)).)))))((....)))))))).............(((..((......))..))). ( -46.80) >DroSec_CAF1 6638 120 - 1 GGCCGCCGAAGAGGCUCGUGUCGCUGAAGAGGCUCGUCUGGAGGAGGAGCAGCGUGUCCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCG ..((((((....(((((.((((((.((.(..((((.((.....)).))))..)...)).)))).)).)))))((....)))))))).............(((..((......))..))). ( -47.80) >DroSim_CAF1 7459 120 - 1 GGCCGCCGAAGAGGCUCGUGUCGCUGAAGAGGCUCGUCUGGAGGAGGAGCAGCGUGUCCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCG ..((((((....(((((.((((((.((.(..((((.((.....)).))))..)...)).)))).)).)))))((....)))))))).............(((..((......))..))). ( -47.80) >DroEre_CAF1 7013 120 - 1 GGCCGCCGAAGAGGCUCGUGUCGCUGAAGAGGCUCGUCUGGAGGAGGAGCAGCGGGUGCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCG ..(((((.....)))(((..((((.......((((.((.....)).))))..(((((((.(.(((....))).).)))))))))))....)))...)).(((..((......))..))). ( -48.80) >DroYak_CAF1 7038 120 - 1 GGCUGCCGAAGAGGCUCGUGUCGCUGAAGAGGCUCGUUUGGAGGAGGAGCAGCGUGUCCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGCGAGGGAGAACUUGCUCG ..((((((....(((((.((((((.((.(..((((.((.....)).))))..)...)).)))).)).)))))((....)))))))).............(((((((......))))))). ( -45.50) >consensus GGCCGCCGAAGAGGCUCGUGUCGCUGAAGAGGCUCGUCUGGAGGAGGAGCAGCGUGUCCGUGAGCAGGAGCUGGAGCGCCUGGCGGAGAUCGAGAAGGAGAGUGAGGGAGAACUUGCUCG ..((((((....(((((.((((((.((.(..((((.((.....)).))))..)...)).)))).)).)))))((....)))))))).............(((((((......))))))). (-45.58 = -45.50 + -0.08)


| Location | 6,594,302 – 6,594,422 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -43.94 |
| Consensus MFE | -41.64 |
| Energy contribution | -41.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6594302 120 + 27905053 UUUUGGGCAGCUUCUUCGGCGGCACGAGCCUCCUCUACAGCCUUCAGUGCGGCUUCUUCGGCGGCCUUUUGGGCAGCUUCUUCAGCCAGACGAGCCUCUUCGGCUGCCUUUAAAGCGGCU .....(((.((((....((((((..(((.(((.(((...(((....).))((((.....(((.(((.....))).))).....))))))).))).)))....))))))....)))).))) ( -43.80) >DroSec_CAF1 6758 120 + 1 UUUUGGGCAGCUUCUUCGGCGGCACGAGCCUCCUCUACAGCCUUCAGUGCGGCUUCUUCGGCGGCCUUUUGGGCAGCUUCUUCAGCCAGACGAGCCUCUUCGGCUGCCUUUAAAGCGGCU .....(((.((((....((((((..(((.(((.(((...(((....).))((((.....(((.(((.....))).))).....))))))).))).)))....))))))....)))).))) ( -43.80) >DroSim_CAF1 7579 119 + 1 U-UUGGGCAGCUUCUUCGGCGGCACGAGCGUCCUCUACAGCCUUCAGUGCGGCUUCUUCGGCGGCCUUUUGGGCAGCUUCUUCAGCCAGACGAGCCUCUUCGGCUGCCUUUAAAGCGGCU .-...(((.((((....((((((.(((((.((.(((...(((....).))((((.....(((.(((.....))).))).....))))))).))))....)))))))))....)))).))) ( -41.40) >DroEre_CAF1 7133 120 + 1 UUUUGGGCAGCUUCUUCGGCGGCACGAGCCUCCUCUACAGCCCUCAGUGCGGCUUCUUCAGCUGCCUUUUGGGCGGCUUCUUCAGCCAGUCGAGCCUCUUCGGCUGCCUUUAGAGCGGCU .....(((.(((.((..((((((.((((....(((.((.((((..((.((((((.....))))))))...))))((((.....)))).)).)))....))))))))))...))))).))) ( -50.70) >DroYak_CAF1 7158 120 + 1 UUUUGGGCAGCUUCUUCGGCGGCACGAGCUUCCUCUACAGCCUUCAGUGCGGCUUCUUCAGCGGCCUUUUGGGCAGCUUCUUCAGCCAGACGAGCCUCUUCGGCGGCCUUUAGAGCGGCU .....(((.(((.((..(((.((..(((.(((.(((...(((....).))((((.....(((.(((.....))).))).....))))))).))).)))....)).)))...))))).))) ( -40.00) >consensus UUUUGGGCAGCUUCUUCGGCGGCACGAGCCUCCUCUACAGCCUUCAGUGCGGCUUCUUCGGCGGCCUUUUGGGCAGCUUCUUCAGCCAGACGAGCCUCUUCGGCUGCCUUUAAAGCGGCU .....(((.((((....((((((..(((.(((.(((...(((....).))((((.....(((.(((.....))).))).....))))))).))).)))....))))))....)))).))) (-41.64 = -41.64 + 0.00)



| Location | 6,594,302 – 6,594,422 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -39.38 |
| Energy contribution | -39.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6594302 120 - 27905053 AGCCGCUUUAAAGGCAGCCGAAGAGGCUCGUCUGGCUGAAGAAGCUGCCCAAAAGGCCGCCGAAGAAGCCGCACUGAAGGCUGUAGAGGAGGCUCGUGCCGCCGAAGAAGCUGCCCAAAA .((.(((((...(((.((....(((.(((.(((((((......((.(((.....))).))......))))((.(....)))...))).))).)))..)).)))...))))).))...... ( -44.10) >DroSec_CAF1 6758 120 - 1 AGCCGCUUUAAAGGCAGCCGAAGAGGCUCGUCUGGCUGAAGAAGCUGCCCAAAAGGCCGCCGAAGAAGCCGCACUGAAGGCUGUAGAGGAGGCUCGUGCCGCCGAAGAAGCUGCCCAAAA .((.(((((...(((.((....(((.(((.(((((((......((.(((.....))).))......))))((.(....)))...))).))).)))..)).)))...))))).))...... ( -44.10) >DroSim_CAF1 7579 119 - 1 AGCCGCUUUAAAGGCAGCCGAAGAGGCUCGUCUGGCUGAAGAAGCUGCCCAAAAGGCCGCCGAAGAAGCCGCACUGAAGGCUGUAGAGGACGCUCGUGCCGCCGAAGAAGCUGCCCAA-A .((.(((((...(((((((.....)))).((.(((((......((.(((.....))).))......))))).))....(((....(((....)))..))))))...))))).))....-. ( -41.80) >DroEre_CAF1 7133 120 - 1 AGCCGCUCUAAAGGCAGCCGAAGAGGCUCGACUGGCUGAAGAAGCCGCCCAAAAGGCAGCUGAAGAAGCCGCACUGAGGGCUGUAGAGGAGGCUCGUGCCGCCGAAGAAGCUGCCCAAAA .((.(((((...(((.((....(((.(((....((((.....))))((((...((((.(((.....))).)).))..)))).......))).)))..)).)))..)).))).))...... ( -46.10) >DroYak_CAF1 7158 120 - 1 AGCCGCUCUAAAGGCCGCCGAAGAGGCUCGUCUGGCUGAAGAAGCUGCCCAAAAGGCCGCUGAAGAAGCCGCACUGAAGGCUGUAGAGGAAGCUCGUGCCGCCGAAGAAGCUGCCCAAAA .((.(((((...(((.(((.....)))..((.(((((.....(((.(((.....))).))).....))))).))....(((....(((....)))..))))))..)).))).))...... ( -42.80) >consensus AGCCGCUUUAAAGGCAGCCGAAGAGGCUCGUCUGGCUGAAGAAGCUGCCCAAAAGGCCGCCGAAGAAGCCGCACUGAAGGCUGUAGAGGAGGCUCGUGCCGCCGAAGAAGCUGCCCAAAA .((.(((((...(((.(((.....)))..((.(((((......((.(((.....))).))......))))).))....(((....(((....)))..))))))...))))).))...... (-39.38 = -39.98 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:27 2006