| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 924,437 – 924,546 |
| Length | 109 |
| Max. P | 0.983110 |

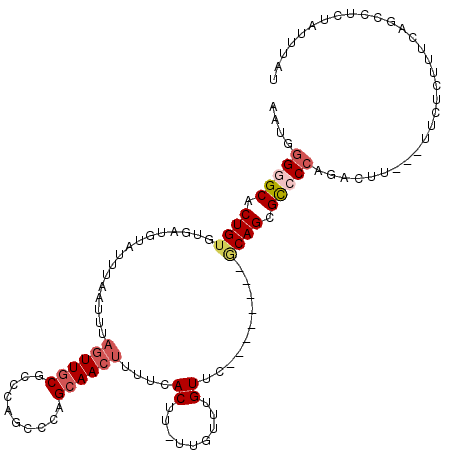
| Location | 924,437 – 924,546 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -17.71 |
| Energy contribution | -18.77 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 924437 109 + 27905053 AUAAAUAGAGGCUGAAAGAGAA---AAGUUUGGGGCACUGU--------GAACAAGCAAAAAGUGAAAAGUUGCUGGGCUGCGUGCAACUAAAUUAAAUACAUCACACAGUGCCCCCAUU ........(((((.........---.)))))((((((((((--------(.....((.....))....((((((..........))))))...............))))))))))).... ( -30.70) >DroSec_CAF1 13422 108 + 1 AUAAAUAGAGGCUGAAAGAGAA---AAGUCUGGGGCGCUGC--------GAACAAACAA-AAGUGAAAAGUUGCUGGGCUGGGCGCAACUAAAUUAAAUACAUCAGACAGUGCCCCCAUU ........(((((.........---.)))))((((((((((--------(..((..(..-.((..(....)..)).)..))..)))..............(....)..)))))))).... ( -28.20) >DroSim_CAF1 19325 108 + 1 AUAAAUAGAGGCUGAAAGAGAA---AAGUCUGGGGCGCUGC--------GAACAAACAA-AAGUGAAAAGUUGCUGGGCUGGGCGCAACUAAAUUAAAUACAUCACACAGUGCCCCCAUU ........(((((.........---.)))))(((((((((.--------(.....((..-..))....((((((..........))))))...............).))))))))).... ( -28.00) >DroEre_CAF1 14522 102 + 1 AUAAAUAGGGGCUGAAAGGGAG---AAGUCUGGUGCGCUGA--------GAACAUAGAG-AAGUGAAAAGUUGCUGG-----GCGCAACUAA-UGAAAUACAUCACACAGUGCCCCCAUU .......(((((.......((.---...(((((((((((.(--------((((......-.........))).)).)-----)))).)))).-.))......)).......))))).... ( -25.70) >DroYak_CAF1 34120 109 + 1 AUAAAUAGAGGCUGAAAAAAAG--UGAGUCUGGGGCGCUGC--------GAACAAACAA-AAGUGAAAAGUUUCUGGGCUGAGCGCAACUAAGUGAAAUACAUCACACAGUGCCCCCAUU ........(((((.(.......--).)))))(((((((((.--------..........-..(((...((((....))))...)))......((((......)))).))))))))).... ( -31.00) >DroAna_CAF1 12121 109 + 1 ACAAAUGCAGGCUGAAGGUACAUAUGGGAGGGGGACACUGUCAAGGGAUGCACAAAUAA-AAGUGAAAAGUUGCAAUUC-CCUUGCAAGA---------ACCUUCAACAGAGCACUCAUA .....(((..(.(((((((.(.........(....)..((.(((((((((((.......-...........))))..))-))))))).).---------))))))).)...)))...... ( -27.27) >consensus AUAAAUAGAGGCUGAAAGAGAA___AAGUCUGGGGCGCUGC________GAACAAACAA_AAGUGAAAAGUUGCUGGGCUGCGCGCAACUAAAUUAAAUACAUCACACAGUGCCCCCAUU ........(((((.............)))))(((((((((...........((.........))....((((((..........)))))).................))))))))).... (-17.71 = -18.77 + 1.06)

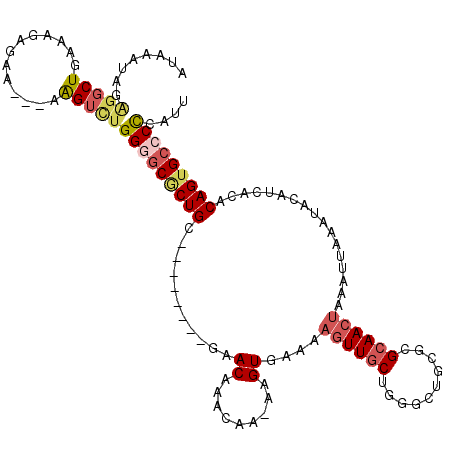
| Location | 924,437 – 924,546 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -27.99 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 924437 109 - 27905053 AAUGGGGGCACUGUGUGAUGUAUUUAAUUUAGUUGCACGCAGCCCAGCAACUUUUCACUUUUUGCUUGUUC--------ACAGUGCCCCAAACUU---UUCUCUUUCAGCCUCUAUUUAU ....(((((((((((....(((........((((((..........))))))..........))).....)--------))))))))))......---...................... ( -31.37) >DroSec_CAF1 13422 108 - 1 AAUGGGGGCACUGUCUGAUGUAUUUAAUUUAGUUGCGCCCAGCCCAGCAACUUUUCACUU-UUGUUUGUUC--------GCAGCGCCCCAGACUU---UUCUCUUUCAGCCUCUAUUUAU (((((((((...(((((..((..........((((((..(((....((((..........-)))))))..)--------)))))))..)))))..---..........)))))))))... ( -22.86) >DroSim_CAF1 19325 108 - 1 AAUGGGGGCACUGUGUGAUGUAUUUAAUUUAGUUGCGCCCAGCCCAGCAACUUUUCACUU-UUGUUUGUUC--------GCAGCGCCCCAGACUU---UUCUCUUUCAGCCUCUAUUUAU ....(((((.(((((.((((..........((((((..........))))))........-.))))....)--------)))).)))))......---...................... ( -26.01) >DroEre_CAF1 14522 102 - 1 AAUGGGGGCACUGUGUGAUGUAUUUCA-UUAGUUGCGC-----CCAGCAACUUUUCACUU-CUCUAUGUUC--------UCAGCGCACCAGACUU---CUCCCUUUCAGCCCCUAUUUAU (((((((((((((.((((......)))-)))))(((((-----..((.(((.........-......))))--------)..)))))........---..........)))))))))... ( -27.26) >DroYak_CAF1 34120 109 - 1 AAUGGGGGCACUGUGUGAUGUAUUUCACUUAGUUGCGCUCAGCCCAGAAACUUUUCACUU-UUGUUUGUUC--------GCAGCGCCCCAGACUCA--CUUUUUUUCAGCCUCUAUUUAU (((((((((((((.((((......)))).)))).(((((..((..(.((((.........-..)))).)..--------)))))))..........--..........)))))))))... ( -28.60) >DroAna_CAF1 12121 109 - 1 UAUGAGUGCUCUGUUGAAGGU---------UCUUGCAAGG-GAAUUGCAACUUUUCACUU-UUAUUUGUGCAUCCCUUGACAGUGUCCCCCUCCCAUAUGUACCUUCAGCCUGCAUUUGU .(..(((((...(((((((((---------.(.(((((((-((..((((.(.........-......)))))))))))).))(((.........)))..).)))))))))..)))))..) ( -31.86) >consensus AAUGGGGGCACUGUGUGAUGUAUUUAAUUUAGUUGCGCCCAGCCCAGCAACUUUUCACUU_UUGUUUGUUC________GCAGCGCCCCAGACUU___UUCUCUUUCAGCCUCUAUUUAU ....(((((.((((................((((((..........))))))....((.........))..........)))).)))))............................... (-14.79 = -15.65 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:17 2006