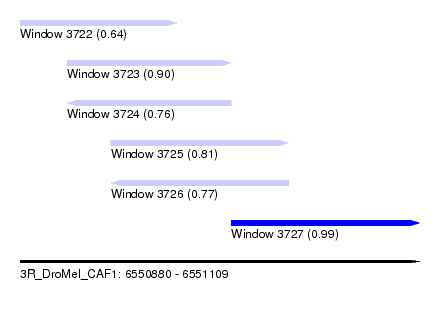
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,550,880 – 6,551,109 |
| Length | 229 |
| Max. P | 0.986783 |

| Location | 6,550,880 – 6,550,970 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.42 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6550880 90 + 27905053 UUUGGCUUCUCUGACAGACAGCCGCAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGAC ((((((((((.....)))..(((........)))........))))))).......((((((....)))))).................. ( -24.30) >DroSec_CAF1 27764 90 + 1 GUUGGCUUCUCUGACAGACAGUCGCAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGAC .((((((..((((((.....)))((.......)).)))....))))))........((((((....)))))).................. ( -23.80) >DroSim_CAF1 27527 90 + 1 UUUGGCUUCUCUGACAGACAGCCGCAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGAC ((((((((((.....)))..(((........)))........))))))).......((((((....)))))).................. ( -24.30) >DroEre_CAF1 30785 90 + 1 UUUGGCUUCUUUGACAGACAGCCGUAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGAC ((((((((((((........(((........))).((.......)).))))))...((((((....)))))).....))))))....... ( -26.20) >DroYak_CAF1 28993 90 + 1 UUUGGCUUCUCUGACAGACAGCCGUAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGAC ((((((((((.....)))..(((........)))........))))))).......((((((....)))))).................. ( -24.30) >consensus UUUGGCUUCUCUGACAGACAGCCGCAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGAC ((((((((((.....)))..(((........)))........))))))).......((((((....)))))).................. (-23.38 = -23.42 + 0.04)

| Location | 6,550,907 – 6,551,001 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6550907 94 + 27905053 UAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUG .....(((((((((((((((((.......((((((....)))))).........(((........)))..))))))))....)))))..)))). ( -31.10) >DroSec_CAF1 27791 94 + 1 UAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUG .....(((((((((((((((((.......((((((....)))))).........(((........)))..))))))))....)))))..)))). ( -31.10) >DroSim_CAF1 27554 94 + 1 UAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUG .....(((((((((((((((((.......((((((....)))))).........(((........)))..))))))))....)))))..)))). ( -31.10) >DroEre_CAF1 30812 94 + 1 UAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUG .....(((((((((((((((((.......((((((....)))))).........(((........)))..))))))))....)))))..)))). ( -31.10) >DroYak_CAF1 29020 94 + 1 UAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUG .....(((((((((((((((((.......((((((....)))))).........(((........)))..))))))))....)))))..)))). ( -31.10) >DroAna_CAF1 33317 76 + 1 ------------------CGAAAGAAAAAGGGCUCAUUUGGGCUCACUCAGAUAGACAUUGACCAGUCCAUUUGGCCAUGUCAAUUUAUCUGCG ------------------(....).....((((((....))))))...(((((((((((.(.((((.....))))).)))))....)))))).. ( -22.60) >consensus UAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUG ....(((.........)))...(((....((((((....))))))....((((.(((((.(.((((.....))))).)))))))))..)))... (-24.20 = -24.48 + 0.28)



| Location | 6,550,907 – 6,551,001 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -21.58 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6550907 94 - 27905053 CACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUUUCUUUUGGCCAAGUUCCUGCCUUUA ..(((.....)))((.(((((((((((((........)))))...(((.((((.(....).))))...))).)))))))).))........... ( -27.20) >DroSec_CAF1 27791 94 - 1 CACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUUUCUUUUGGCCAAGUUCCUGCCUUUA ..(((.....)))((.(((((((((((((........)))))...(((.((((.(....).))))...))).)))))))).))........... ( -27.20) >DroSim_CAF1 27554 94 - 1 CACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUUUCUUUUGGCCAAGUUCCUGCCUUUA ..(((.....)))((.(((((((((((((........)))))...(((.((((.(....).))))...))).)))))))).))........... ( -27.20) >DroEre_CAF1 30812 94 - 1 CACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUUUCUUUUGGCCAAGUUCCUGCCUUUA ..(((.....)))((.(((((((((((((........)))))...(((.((((.(....).))))...))).)))))))).))........... ( -27.20) >DroYak_CAF1 29020 94 - 1 CACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUUUCUUUUGGCCAAGUUCCUGCCUUUA ..(((.....)))((.(((((((((((((........)))))...(((.((((.(....).))))...))).)))))))).))........... ( -27.20) >DroAna_CAF1 33317 76 - 1 CGCAGAUAAAUUGACAUGGCCAAAUGGACUGGUCAAUGUCUAUCUGAGUGAGCCCAAAUGAGCCCUUUUUCUUUCG------------------ ..((((((....((((((((((.......))))).)))))))))))((.(.((.(....).)))))..........------------------ ( -16.70) >consensus CACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUUUCUUUUGGCCAAGUUCCUGCCUUUA ..(((.....)))...(((((((((((((........)))))...(((.((((.(....).))))...))).)))))))).............. (-21.58 = -22.30 + 0.72)

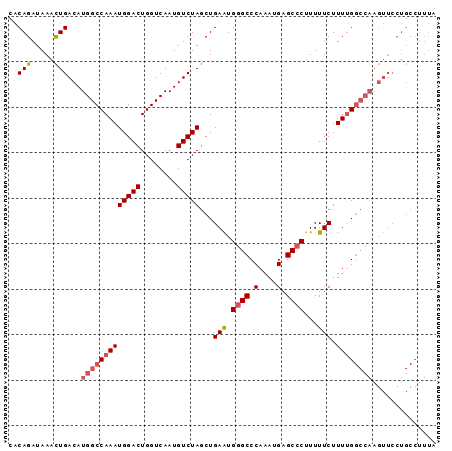
| Location | 6,550,932 – 6,551,034 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 96.87 |
| Mean single sequence MFE | -30.99 |
| Consensus MFE | -25.44 |
| Energy contribution | -25.19 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
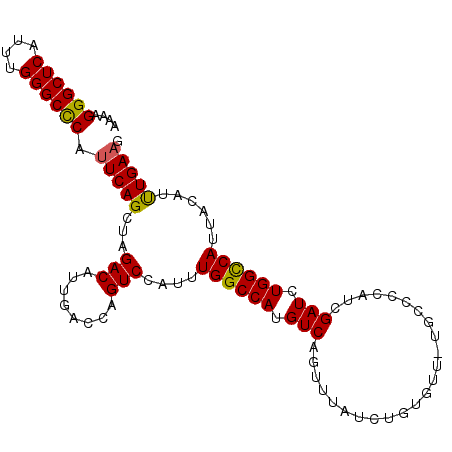
>3R_DroMel_CAF1 6550932 102 + 27905053 AAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUGUU-UGCCCCAUCGAUCUGGUCAUUACAUUUGAAG ....((((((....)))))).(((((........(((((((((.((..(((.....(((.....)))....-.)))..)).)).)))))))......))))). ( -31.64) >DroSec_CAF1 27816 102 + 1 AAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUGUU-UGCCCCAUCGAUCUGGCCAUUACAUUUGAAG ....((((((....)))))).(((((((.(((((.(.((((.....))))).)))))))).....((((..-.(((..........)))...))))..)))). ( -28.70) >DroSim_CAF1 27579 102 + 1 AAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUGUU-UGGCCCAUCGAUCUGGCCAUUACAUUUGAAG ....((((((....)))))).(((((((.(((((.(.((((.....))))).)))))))).....((((..-(((((.........))))).))))..)))). ( -33.50) >DroEre_CAF1 30837 102 + 1 AAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUGUU-UGCCCCAUCGAUCUGGCCAUUACAUUUGAAG ....((((((....)))))).(((((((.(((((.(.((((.....))))).)))))))).....((((..-.(((..........)))...))))..)))). ( -28.70) >DroYak_CAF1 29045 102 + 1 AAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUGUU-UGCCCCAUCGAUCUGGCCAUUACAUUUGAAG ....((((((....)))))).(((((((.(((((.(.((((.....))))).)))))))).....((((..-.(((..........)))...))))..)))). ( -28.70) >DroAna_CAF1 33324 103 + 1 AAAAGGGCUCAUUUGGGCUCACUCAGAUAGACAUUGACCAGUCCAUUUGGCCAUGUCAAUUUAUCUGCGUUUUGGCCCAUCGAUCUGGCCAUUACAUCUGAAG ....((((((....))))))..((((((.(((((.(.((((.....))))).)))))...........((..(((((.........)))))..)))))))).. ( -34.70) >consensus AAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAUGUCAGUUUAUCUGUGUU_UGCCCCAUCGAUCUGGCCAUUACAUUUGAAG ....((((((....)))))).(((((...(((........)))....((((((.(((........................))).))))))......))))). (-25.44 = -25.19 + -0.25)
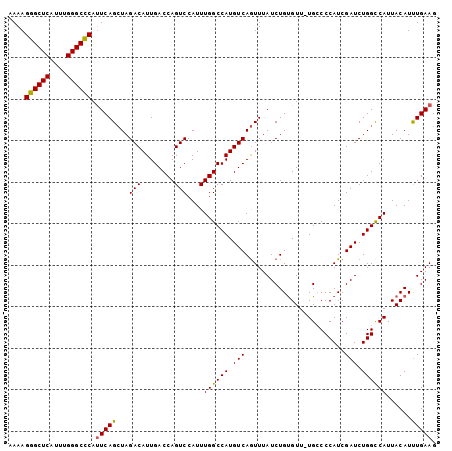
| Location | 6,550,932 – 6,551,034 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 96.87 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -25.25 |
| Energy contribution | -25.33 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6550932 102 - 27905053 CUUCAAAUGUAAUGACCAGAUCGAUGGGGCA-AACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUU .((((...(((.(((((((.((.((..(((.-....(((.....))).....)))..)).))))))))).)).)....)))).((((.(....).)))).... ( -30.10) >DroSec_CAF1 27816 102 - 1 CUUCAAAUGUAAUGGCCAGAUCGAUGGGGCA-AACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUU .((((...(((.(((((((.((.((..(((.-....(((.....))).....)))..)).))))))))).)).)....)))).((((.(....).)))).... ( -29.50) >DroSim_CAF1 27579 102 - 1 CUUCAAAUGUAAUGGCCAGAUCGAUGGGCCA-AACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUU .((((...(((.(((((((.((.((.(((((-....(((.....)))...)))))..)).))))))))).)).)....)))).((((.(....).)))).... ( -30.40) >DroEre_CAF1 30837 102 - 1 CUUCAAAUGUAAUGGCCAGAUCGAUGGGGCA-AACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUU .((((...(((.(((((((.((.((..(((.-....(((.....))).....)))..)).))))))))).)).)....)))).((((.(....).)))).... ( -29.50) >DroYak_CAF1 29045 102 - 1 CUUCAAAUGUAAUGGCCAGAUCGAUGGGGCA-AACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUU .((((...(((.(((((((.((.((..(((.-....(((.....))).....)))..)).))))))))).)).)....)))).((((.(....).)))).... ( -29.50) >DroAna_CAF1 33324 103 - 1 CUUCAGAUGUAAUGGCCAGAUCGAUGGGCCAAAACGCAGAUAAAUUGACAUGGCCAAAUGGACUGGUCAAUGUCUAUCUGAGUGAGCCCAAAUGAGCCCUUUU .............(((((......(((((....((.((((((....((((((((((.......))))).))))))))))).))..)))))..)).)))..... ( -31.30) >consensus CUUCAAAUGUAAUGGCCAGAUCGAUGGGGCA_AACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCUAGCUGAAUGGGCCCAAAUGAGCCCUUUU .((((...(((.(((((((.((.((..(((......(((.....))).....)))..)).))))))))).)).)....)))).((((.(....).)))).... (-25.25 = -25.33 + 0.09)


| Location | 6,551,001 – 6,551,109 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.49 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -27.18 |
| Energy contribution | -26.70 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6551001 108 + 27905053 UUUGCCCCAUCGAUCUGGUCAUUACAUUUGAAGUGUUAUGGCUUGGACAUCAGCACGUAACAUGGGUUCUUUUUGGCCAACUUCGACUUCGGAUAUGGUUAUCUGCCU--- .........((((..((((((...........(((((((((((........))).))))))))..........))))))...))))...((((((....))))))...--- ( -27.10) >DroSec_CAF1 27885 108 + 1 UUUGCCCCAUCGAUCUGGCCAUUACAUUUGAAGUGUUAUGGCUUGGACAUCAGCACGUAACACGGGUUCUUUUUGGCCAACUUCGACUUGGGCUAUGGCUAUCUGCCU--- ...((((..((((..((((((...........(((((((((((........))).))))))))..........))))))...))))...))))...(((.....))).--- ( -39.20) >DroSim_CAF1 27648 108 + 1 UUUGGCCCAUCGAUCUGGCCAUUACAUUUGAAGUGUUAUGGCUUGGACAUCAGCACGUAACACGGGUUCUUUUUGGCCAACUUCGACUUGGGCUAUAGCUAUCUGCCU--- ..(((((((((((..((((((...........(((((((((((........))).))))))))..........))))))...))))..)))))))..((.....))..--- ( -39.20) >DroEre_CAF1 30906 102 + 1 UUUGCCCCAUCGAUCUGGCCAUUACAUUUGAAGUGUUAUGGCUUGGACAUCAGCACGUAACACGGGUUCUUUUUGGCCAACUUCGAC------UAUGGCUAUCUGUCU--- ...(((...((((..((((((...........(((((((((((........))).))))))))..........))))))...)))).------...))).........--- ( -31.20) >DroYak_CAF1 29114 111 + 1 UUUGCCCCAUCGAUCUGGCCAUUACAUUUGAAGUGUUAUGGCUUGGACAUCAGCACGUAACACGGGUUCUUUUUGGCAAACUUCGGCAUCGGGUAUGGCUAUCUGUCUAUC ..(((((..((((....((((...........(((((((((((........))).))))))))..........)))).....))))....))))).(((.....))).... ( -29.90) >consensus UUUGCCCCAUCGAUCUGGCCAUUACAUUUGAAGUGUUAUGGCUUGGACAUCAGCACGUAACACGGGUUCUUUUUGGCCAACUUCGACUUCGGCUAUGGCUAUCUGCCU___ .........((((..((((((...........(((((((((((........))).))))))))..........))))))...))))..........(((.....))).... (-27.18 = -26.70 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:07 2006