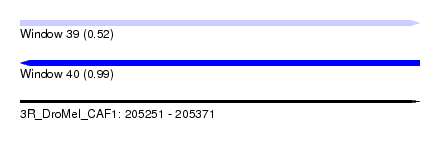
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 205,251 – 205,371 |
| Length | 120 |
| Max. P | 0.993958 |

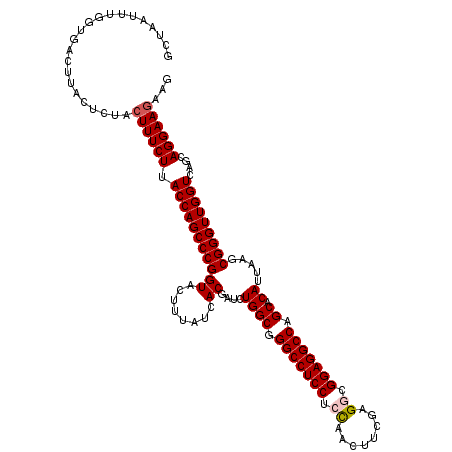
| Location | 205,251 – 205,371 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -33.56 |
| Energy contribution | -34.28 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 205251 120 + 27905053 CUUCUUCCUGCUGACCAACCCGCUUAAUGUGCUGGCCUCCGCCUCGAAGUUGGAGGAGGCCCGCCAGAUUGUGAUAAAGUACCGGGCUGGUAAGAAAGAAGAGUAGGUAACCAAAUUAGA .((((((...((.((((.((((......((((((((((((.((........)).)))))))(((......)))....))))))))).)))).))...))))))................. ( -42.00) >DroSec_CAF1 30998 120 + 1 CUUCUUCCUGCUGACCAACCCGCUGAAUGUGCUGGCCUCCGCCUCGAAGUUGGAGGAGGCCCGCCAGAUCGUGAUAAAGUACCGGGCUGGUAAGAAAGUAGAGUAAGUCACCAAAUUAGC ...(((((((((.((((.((((..((.((.((.(((((((.((........)).))))))).))))..))(((......))))))).)))).....))))).).)))............. ( -40.90) >DroSim_CAF1 31788 120 + 1 CUUCUUCCUGCUGACCAACCCGCUGAAUGUGCUGGCCUCCGCCUCGAAGUUGGAGGAGGCCCGCCAGAUCGUGAUAAAGUACCGGGCUGGUAAGAAAGUAGAGUAAGUCACCAAAUUAGC ...(((((((((.((((.((((..((.((.((.(((((((.((........)).))))))).))))..))(((......))))))).)))).....))))).).)))............. ( -40.90) >DroEre_CAF1 32920 118 + 1 CUUCUUCCUGCUGACCAACCCGCUUAAUGUGCUGGCCUCCGACUCGAAGUUGGAGGAGGCCCGCCAGAUAGUGCUGAGGUACCGGGCUGGUAAGAAAC--CAAUUAGUCACCAAUUUAGC .(((((((.(((.(((....((((...((.((.(((((((..(........)..))))))).))))...))))....)))....))).)).)))))..--.................... ( -38.20) >DroYak_CAF1 32636 120 + 1 CUUCUUCCUGCUGACCAACCCGCUUAAUGUGCUGGCCUCCGACUCGAAGUUAGAGGAGGCCCGCCAGAUAGUGCUGAGGUACCGGGCUGGUAAGAAAGAGGACUCUAUCACCAAUUUAGC ((((((.((....((((.((((.....((.((.((((((((((.....)))...))))))).))))....((((....)))))))).)))).)).))))))................... ( -41.10) >consensus CUUCUUCCUGCUGACCAACCCGCUUAAUGUGCUGGCCUCCGCCUCGAAGUUGGAGGAGGCCCGCCAGAUAGUGAUAAAGUACCGGGCUGGUAAGAAAGAAGAGUAAGUCACCAAAUUAGC .((((.....((.((((.((((......((((((((((((.((........)).)))))))(((......)))....))))))))).)))).)).....))))................. (-33.56 = -34.28 + 0.72)

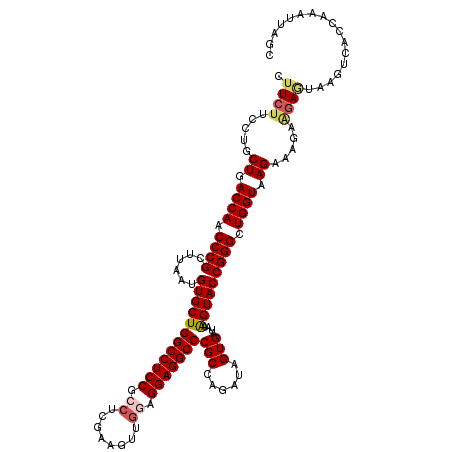
| Location | 205,251 – 205,371 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -44.04 |
| Consensus MFE | -38.20 |
| Energy contribution | -38.64 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 205251 120 - 27905053 UCUAAUUUGGUUACCUACUCUUCUUUCUUACCAGCCCGGUACUUUAUCACAAUCUGGCGGGCCUCCUCCAACUUCGAGGCGGAGGCCAGCACAUUAAGCGGGUUGGUCAGCAGGAAGAAG ..................(((((((.((.(((((((((((........))....((((.(((((((.((........)).))))))).)).)).....))))))))).)).))))))).. ( -47.20) >DroSec_CAF1 30998 120 - 1 GCUAAUUUGGUGACUUACUCUACUUUCUUACCAGCCCGGUACUUUAUCACGAUCUGGCGGGCCUCCUCCAACUUCGAGGCGGAGGCCAGCACAUUCAGCGGGUUGGUCAGCAGGAAGAAG ........(....)....(((.(((.((.(((((((((((........))((....((.(((((((.((........)).))))))).))....))..))))))))).)).))).))).. ( -45.20) >DroSim_CAF1 31788 120 - 1 GCUAAUUUGGUGACUUACUCUACUUUCUUACCAGCCCGGUACUUUAUCACGAUCUGGCGGGCCUCCUCCAACUUCGAGGCGGAGGCCAGCACAUUCAGCGGGUUGGUCAGCAGGAAGAAG ........(....)....(((.(((.((.(((((((((((........))((....((.(((((((.((........)).))))))).))....))..))))))))).)).))).))).. ( -45.20) >DroEre_CAF1 32920 118 - 1 GCUAAAUUGGUGACUAAUUG--GUUUCUUACCAGCCCGGUACCUCAGCACUAUCUGGCGGGCCUCCUCCAACUUCGAGUCGGAGGCCAGCACAUUAAGCGGGUUGGUCAGCAGGAAGAAG (((((((..((.....))..--))))...(((((((((.(....(((......)))((.(((((((..(........)..))))))).))......).))))))))).)))......... ( -41.50) >DroYak_CAF1 32636 120 - 1 GCUAAAUUGGUGAUAGAGUCCUCUUUCUUACCAGCCCGGUACCUCAGCACUAUCUGGCGGGCCUCCUCUAACUUCGAGUCGGAGGCCAGCACAUUAAGCGGGUUGGUCAGCAGGAAGAAG ..................((.((((.((.(((((((((.(....(((......)))((.(((((((.((.......))..))))))).))......).))))))))).)).)))).)).. ( -41.10) >consensus GCUAAUUUGGUGACUUACUCUACUUUCUUACCAGCCCGGUACUUUAUCACGAUCUGGCGGGCCUCCUCCAACUUCGAGGCGGAGGCCAGCACAUUAAGCGGGUUGGUCAGCAGGAAGAAG ......................((((((.(((((((((((........))....((((.(((((((.((........)).))))))).)).)).....)))))))))....))))))... (-38.20 = -38.64 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:31 2006