
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,522,274 – 6,522,427 |
| Length | 153 |
| Max. P | 0.926865 |

| Location | 6,522,274 – 6,522,394 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.11 |
| Mean single sequence MFE | -23.31 |
| Consensus MFE | -13.94 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6522274 120 + 27905053 UUGGAAAGAAAAGCUGGCAAAUAGCUGACCAAUAUUUCGCUACAGCGUUUCAUACCAAAUGGGAACACACCCUUGUUAAUAGUAUCGAAUAACUAUCGACUGCUGUUCCACCUCCAGCAA (((((..((((.((((.....((((.((........)))))))))).))))..........((((((((.....((..(((((........)))))..)))).))))))...)))))... ( -27.60) >DroSec_CAF1 7408 101 + 1 ----AAAAAAAACCUAGCAAAUAGCUGACCAAUAUC-------------UCAUCUCCAAUGGGAAC--ACCCUUGUUAAUAGUAUCGAAUAACUAUCGACUGCUGUUCCACCUCCAGCAA ----...................((((.........-------------.(((.....)))(((((--(.(...((..(((((........)))))..)).).)))))).....)))).. ( -19.30) >DroSim_CAF1 8786 105 + 1 UUGUAAAGAAAACCUGGAAAAUAGCUGACCAAUAUC-------------UCAUCUCCAAUGGGAAC--ACCCUUGUUAAUAGUAUCGAAUAACUAUCGACUGCCCUUCCACCUCCAGCAA .............(((((................((-------------((((.....))))))..--......((..(((((........)))))..))............)))))... ( -16.30) >DroEre_CAF1 7374 118 + 1 UGGUGGAGAAUAGACAGCAAACUGCUGACAAAGUUUUAGCAACAGCAGUUUAUGCAUAAUGGGAAC--ACCCUUGUUAAUAAUAUCGAAUAACUAUCGACGAUUGUUCCACCCCCAGCAA .((((((.....(((((((((((((((...............))))))))..))).....(((...--.))).)))).(((((.((((.......))))..)))))))))))........ ( -30.76) >DroYak_CAF1 7552 115 + 1 UUCUGGAAAACAGCUAGCAAACAGCUGAUCAAAUUUUAGCAAGCGC---UCAUCCACAAUAGGAAC--ACCCUUACUAAUAGCAUCGAACAACUAUCGACUGCAGUUCCACCGCCAGCAA ............(((.((.....(((((.......)))))......---............(((((--.....((....))(((((((.......)))).))).)))))...)).))).. ( -22.60) >consensus UUGUAAAGAAAAGCUAGCAAAUAGCUGACCAAUAUUU_GC_A__GC___UCAUCCCCAAUGGGAAC__ACCCUUGUUAAUAGUAUCGAAUAACUAUCGACUGCUGUUCCACCUCCAGCAA .......................((((.................................(((......))).....(((((((((((.......)))).))))))).......)))).. (-13.94 = -14.30 + 0.36)



| Location | 6,522,274 – 6,522,394 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.11 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -16.28 |
| Energy contribution | -15.48 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6522274 120 - 27905053 UUGCUGGAGGUGGAACAGCAGUCGAUAGUUAUUCGAUACUAUUAACAAGGGUGUGUUCCCAUUUGGUAUGAAACGCUGUAGCGAAAUAUUGGUCAGCUAUUUGCCAGCUUUUCUUUCCAA ....(((((..((((.(((.((..(((((........)))))..))..(((......)))...(((((......((((.(.(((....))).)))))....)))))))).))))))))). ( -32.70) >DroSec_CAF1 7408 101 - 1 UUGCUGGAGGUGGAACAGCAGUCGAUAGUUAUUCGAUACUAUUAACAAGGGU--GUUCCCAUUGGAGAUGA-------------GAUAUUGGUCAGCUAUUUGCUAGGUUUUUUUU---- ..((((.........))))(((.(((((((..(((((((((((..((((((.--...))).)))..))).)-------------).))))))..))))))).)))...........---- ( -26.60) >DroSim_CAF1 8786 105 - 1 UUGCUGGAGGUGGAAGGGCAGUCGAUAGUUAUUCGAUACUAUUAACAAGGGU--GUUCCCAUUGGAGAUGA-------------GAUAUUGGUCAGCUAUUUUCCAGGUUUUCUUUACAA ...((((((((((...(((.(((((.......)))))..((((..((((((.--...))).)))..)))).-------------.......)))..))).)))))))............. ( -26.00) >DroEre_CAF1 7374 118 - 1 UUGCUGGGGGUGGAACAAUCGUCGAUAGUUAUUCGAUAUUAUUAACAAGGGU--GUUCCCAUUAUGCAUAAACUGCUGUUGCUAAAACUUUGUCAGCAGUUUGCUGUCUAUUCUCCACCA ........((((((......(((((.......)))))...........(((.--...)))..((.(((((((((((((..((.........)))))))))))).))).))...)))))). ( -32.80) >DroYak_CAF1 7552 115 - 1 UUGCUGGCGGUGGAACUGCAGUCGAUAGUUGUUCGAUGCUAUUAGUAAGGGU--GUUCCUAUUGUGGAUGA---GCGCUUGCUAAAAUUUGAUCAGCUGUUUGCUAGCUGUUUUCCAGAA ...((((....(((((.(((((.((((((((.((((.....((((((((.((--..(((......)))...---)).))))))))...)))).)))))))).))).)).))))))))).. ( -40.30) >consensus UUGCUGGAGGUGGAACAGCAGUCGAUAGUUAUUCGAUACUAUUAACAAGGGU__GUUCCCAUUGGGGAUGA___GC__U_GC_AAAUAUUGGUCAGCUAUUUGCUAGCUUUUCUUUACAA ..(((((.............((..(((((........)))))..))..(((......)))................................)))))....................... (-16.28 = -15.48 + -0.80)

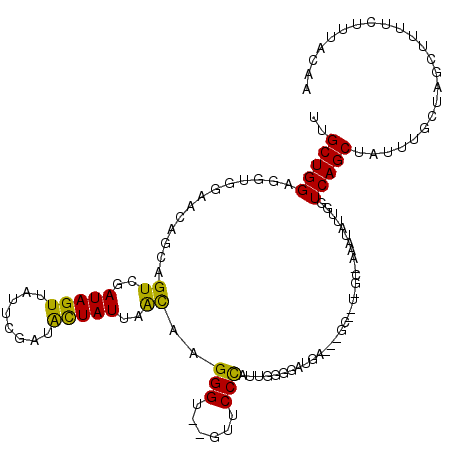
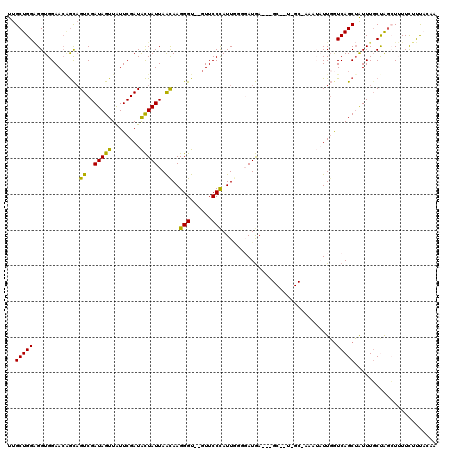
| Location | 6,522,314 – 6,522,427 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6522314 113 + 27905053 UACAGCGUUUCAUACCAAAUGGGAACACACCCUUGUUAAUAGUAUCGAAUAACUAUCGACUGCUGUUCCACCUCCAGCAAAAACGUGUGUUUUUGUUGCGCUGCUAUUUGUUU ..((((((..((..((....))((((((((....((..(((((........)))))..))(((((.........))))).....)))))))).))..)))))).......... ( -31.90) >DroSec_CAF1 7440 102 + 1 ---------UCAUCUCCAAUGGGAAC--ACCCUUGUUAAUAGUAUCGAAUAACUAUCGACUGCUGUUCCACCUCCAGCAAAAACGUGUGUUUUUGUUGCGCUGCUAUUUGUUU ---------............(((((--(.(...((..(((((........)))))..)).).)))))).....((((((((((....))))))))))............... ( -23.10) >DroSim_CAF1 8822 102 + 1 ---------UCAUCUCCAAUGGGAAC--ACCCUUGUUAAUAGUAUCGAAUAACUAUCGACUGCCCUUCCACCUCCAGCAAAAACGUGUGUUUUUGUUGCGCUGCUAUUUGUUU ---------.......(((.(((...--.))))))..(((((((((((.......))))...............((((((((((....))))))))))...)))))))..... ( -19.80) >DroEre_CAF1 7414 99 + 1 AACAGCAGUUUAUGCAUAAUGGGAAC--ACCCUUGUUAAUAAUAUCGAAUAACUAUCGACGAUUGUUCCACCCCCAGCAAGAACGUGUGUUGCUGCUGAGC------------ ..(((((((..((((((...((....--.))((((((((((((.((((.......))))..))))))........))))))...)))))).)))))))...------------ ( -24.80) >DroYak_CAF1 7592 108 + 1 AAGCGC---UCAUCCACAAUAGGAAC--ACCCUUACUAAUAGCAUCGAACAACUAUCGACUGCAGUUCCACCGCCAGCAAGAACGUGUGUUUUUGUUGCGCUGCUAUUUGUUU .(((..---............(((((--.....((....))(((((((.......)))).))).)))))...((((((((((((....)))))))))).)).)))........ ( -26.80) >consensus _A__GC___UCAUCCCCAAUGGGAAC__ACCCUUGUUAAUAGUAUCGAAUAACUAUCGACUGCUGUUCCACCUCCAGCAAAAACGUGUGUUUUUGUUGCGCUGCUAUUUGUUU ....................(((......))).....(((((((((((.......)))).))))))).......((((((((((....))))))))))............... (-16.96 = -17.44 + 0.48)

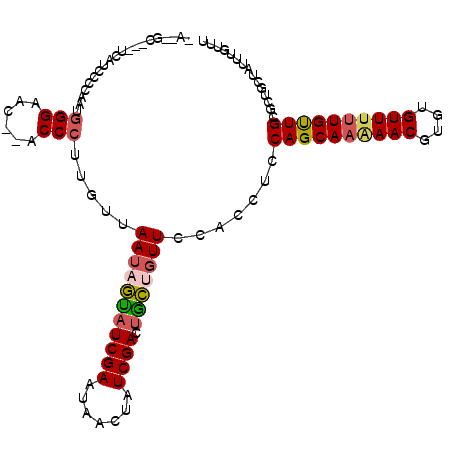

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:52 2006