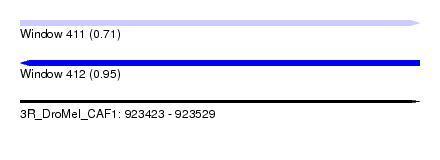
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 923,423 – 923,529 |
| Length | 106 |
| Max. P | 0.953294 |

| Location | 923,423 – 923,529 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.37 |
| Mean single sequence MFE | -15.63 |
| Consensus MFE | -2.58 |
| Energy contribution | -2.38 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.17 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 923423 106 + 27905053 CAUGAUAAUUGUCUAAACC------UUUCAUUCAACCACCUUCACCGGAAAUUCUUCAAACUCCACUUGUGAAAUUCAU----CAACUGCUU---UUUUCUACAAUCAUGAAAGGCUU-C .................((------((((((...............(((............)))..((((((((.....----.........---.)))).))))..))))))))...-. ( -12.56) >DroSec_CAF1 12415 113 + 1 CAUGAUAAUUGUCUAAACC------UUUCAUUCAACCACCUUCACCGGAAACUCUUCAAACUCCCCUUAUGAACUUUAUCGUUCAACUGUUUUUUUUUUCUACAAUCAUGAAGGGCUU-C .................((------((((((...............(....).................(((((......)))))......................))))))))...-. ( -14.20) >DroSim_CAF1 18306 113 + 1 CAUGAUAAUUGUCCAAACC------UUUCAUUCAACCACCUUCACCGGAAACUCUUCAAACACCACUUAUGAACUUUAUCGUUCAACUGUUUUUUUUUUCUACAAUCAUGAAGGGCUU-C .................((------((((((...............(....).................(((((......)))))......................))))))))...-. ( -14.20) >DroEre_CAF1 13624 97 + 1 UAUAAUAAAUGCCUAAACC------UUUCAUUUAAGCUCCGUCACUGUAAACUCUUCAGACUUCACUUGUGAAAUUUAGAGUUCAACGGUUUUUU----------------ACAGCUU-C .....((((((........------...))))))((((.....(((((.((((((..(((.((((....)))).)))))))))..))))).....----------------..)))).-. ( -14.90) >DroYak_CAF1 32594 106 + 1 ------AGAUGCCUAAACUUACAAUUUUCAUUCAACCUCCGUCACCGGAAAUUC-UCAGAUUUCAAUUAUGAAAUUUAGAGUUAAACUGCUUUUAUU-------GUCAAGAAGGGCUUUA ------....((((...((((((((............((((....))))(((((-(.((((((((....))))))))))))))...........)))-------)).)))..)))).... ( -22.30) >consensus CAUGAUAAUUGUCUAAACC______UUUCAUUCAACCACCUUCACCGGAAACUCUUCAAACUCCACUUAUGAAAUUUAUAGUUCAACUGUUUUUUUUUUCUACAAUCAUGAAGGGCUU_C ..........((((.....................((.........))......((((...........)))).......................................)))).... ( -2.58 = -2.38 + -0.20)

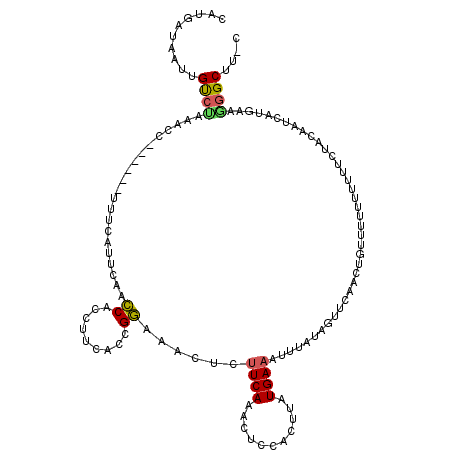
| Location | 923,423 – 923,529 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.37 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -5.26 |
| Energy contribution | -6.86 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.22 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 923423 106 - 27905053 G-AAGCCUUUCAUGAUUGUAGAAAA---AAGCAGUUG----AUGAAUUUCACAAGUGGAGUUUGAAGAAUUUCCGGUGAAGGUGGUUGAAUGAAA------GGUUUAGACAAUUAUCAUG .-(((((((((((((((((......---..))))))(----((.(.((((((....(((((((...)))))))..)))))).).)))..))))))------))))).............. ( -25.40) >DroSec_CAF1 12415 113 - 1 G-AAGCCCUUCAUGAUUGUAGAAAAAAAAAACAGUUGAACGAUAAAGUUCAUAAGGGGAGUUUGAAGAGUUUCCGGUGAAGGUGGUUGAAUGAAA------GGUUUAGACAAUUAUCAUG (-((((.(((((.((((((...........))))))((((......))))............))))).))))).......((((((((..(((..------...)))..))))))))... ( -22.80) >DroSim_CAF1 18306 113 - 1 G-AAGCCCUUCAUGAUUGUAGAAAAAAAAAACAGUUGAACGAUAAAGUUCAUAAGUGGUGUUUGAAGAGUUUCCGGUGAAGGUGGUUGAAUGAAA------GGUUUGGACAAUUAUCAUG (-((((.(((((.((((((...........))))))((((......))))............))))).)))))..((((..((.(((.((.....------...)).))).))..)))). ( -22.10) >DroEre_CAF1 13624 97 - 1 G-AAGCUGU----------------AAAAAACCGUUGAACUCUAAAUUUCACAAGUGAAGUCUGAAGAGUUUACAGUGACGGAGCUUAAAUGAAA------GGUUUAGGCAUUUAUUAUA .-.....((----------------(((...((((((((((((..((((((....))))))....))))))).....))))).((((((((....------.)))))))).))))).... ( -23.20) >DroYak_CAF1 32594 106 - 1 UAAAGCCCUUCUUGAC-------AAUAAAAGCAGUUUAACUCUAAAUUUCAUAAUUGAAAUCUGA-GAAUUUCCGGUGACGGAGGUUGAAUGAAAAUUGUAAGUUUAGGCAUCU------ ....(((...((((.(-------(((.............(((...((((((....))))))..))-)((((((((....))))))))........))))))))....)))....------ ( -27.90) >consensus G_AAGCCCUUCAUGAUUGUAGAAAAAAAAAACAGUUGAACGAUAAAUUUCAUAAGUGGAGUUUGAAGAGUUUCCGGUGAAGGUGGUUGAAUGAAA______GGUUUAGACAAUUAUCAUG ....(((...................................(((((((((....)))))))))..(......))))...((((((((..((((.........))))..))))))))... ( -5.26 = -6.86 + 1.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:14 2006