
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
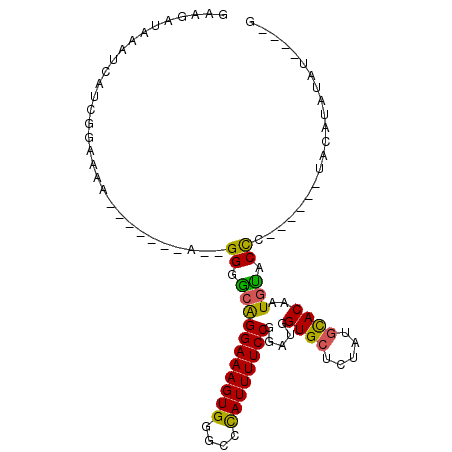
| Location | 6,505,656 – 6,505,748 |
| Length | 92 |
| Max. P | 0.885773 |

| Location | 6,505,656 – 6,505,748 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.00 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
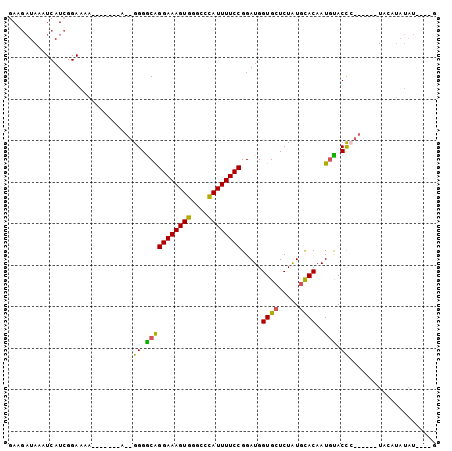
>3R_DroMel_CAF1 6505656 92 + 27905053 GAAGAUAAAUCAUCGGAAAAGAAAAAGA--GGGGCGGGAAAGUGGGCCCAUUUUCCGGAUGGUGCUCUAUGCACAAUUUACCC-----GUAGAUAUAU----G ....(((.(((.((......))......--(((.(.((((((((....)))))))).)...((((.....))))......)))-----...))).)))----. ( -25.00) >DroSec_CAF1 270668 83 + 1 GAAGAUAAAUCAUCGGAA-A-------A--GGGACAGGAAAGUGGGCCUAUUUUCCGGAUGGUGGUCUGUGCACAAUGUACCC------UACAUAUAU----G ..((((..((((((((((-(-------(--(((.((......))..))).)))))).)))))).))))((((.....))))..------.........----. ( -20.10) >DroSim_CAF1 299678 88 + 1 GAAGAUAAAUCAUCGGAAAA-------A--GGGGCAGGAAAGUGGGCCUAUUUUCCGGAUGGUGCUCUAUGCACAAUGUACCC------UACAUAUAUGUAUG ..(((...((((((((((((-------.--(((.(.........).))).)))))).))))))..)))(((((..(((((...------)))))...))))). ( -22.60) >DroEre_CAF1 283891 82 + 1 UA-ACUAAAUCAUCGGAAAA-------GUGGGGGCAGGAAAGUGGGGCCAUUUUCCGGAUGGUGCUCUAUGCACAAUGCACCAUAUAUGU------------- ..-.................-------((((..(((((((((((....)))))))).....((((.....))))..))).))))......------------- ( -25.00) >DroYak_CAF1 279933 93 + 1 GA-GAUAAAUCAUCGGAAAA-------A--GGGGCAGGAAAGUGGGGCCAUUUUCCGGAUGGUGCUCUAUGUACAAUGCACUAUAUACAUACAUAUAUACAUA ((-(....((((((((((((-------.--..(((...........))).)))))).)))))).)))((((((........(((((......))))))))))) ( -20.20) >consensus GAAGAUAAAUCAUCGGAAAA_______A__GGGGCAGGAAAGUGGGCCCAUUUUCCGGAUGGUGCUCUAUGCACAAUGUACCC______UACAUAUAU____G ..............................((.(((((((((((....)))))))).....((((.....))))..))).))..................... (-17.72 = -17.00 + -0.72)

| Location | 6,505,656 – 6,505,748 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -8.67 |
| Energy contribution | -9.27 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
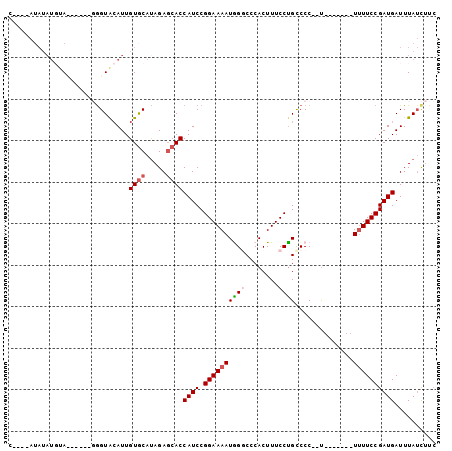
>3R_DroMel_CAF1 6505656 92 - 27905053 C----AUAUAUCUAC-----GGGUAAAUUGUGCAUAGAGCACCAUCCGGAAAAUGGGCCCACUUUCCCGCCCC--UCUUUUUCUUUUCCGAUGAUUUAUCUUC .----..........-----((((((((.((((.....))))((((.((((((.((((..........)))).--........)))))))))))))))))).. ( -21.10) >DroSec_CAF1 270668 83 - 1 C----AUAUAUGUA------GGGUACAUUGUGCACAGACCACCAUCCGGAAAAUAGGCCCACUUUCCUGUCCC--U-------U-UUCCGAUGAUUUAUCUUC (----(((.(((((------...)))))))))..........((((.(((((..(((..((......))..))--)-------)-)))))))).......... ( -14.10) >DroSim_CAF1 299678 88 - 1 CAUACAUAUAUGUA------GGGUACAUUGUGCAUAGAGCACCAUCCGGAAAAUAGGCCCACUUUCCUGCCCC--U-------UUUUCCGAUGAUUUAUCUUC ..((((....))))------(((((....((((.....))))((((.((((((..(((..........)))..--.-------))))))))))...))))).. ( -19.80) >DroEre_CAF1 283891 82 - 1 -------------ACAUAUAUGGUGCAUUGUGCAUAGAGCACCAUCCGGAAAAUGGCCCCACUUUCCUGCCCCCAC-------UUUUCCGAUGAUUUAGU-UA -------------.(((..(((((((.(((....))).))))))).((((((((((...((......))...))).-------)))))))))).......-.. ( -19.80) >DroYak_CAF1 279933 93 - 1 UAUGUAUAUAUGUAUGUAUAUAGUGCAUUGUACAUAGAGCACCAUCCGGAAAAUGGCCCCACUUUCCUGCCCC--U-------UUUUCCGAUGAUUUAUC-UC ((((((((.((((((.......))))))))))))))......((((.((((((.(((...........)))..--.-------)))))))))).......-.. ( -23.00) >consensus C____AUAUAUGUA______GGGUACAUUGUGCAUAGAGCACCAUCCGGAAAAUGGGCCCACUUUCCUGCCCC__U_______UUUUCCGAUGAUUUAUCUUC .............................((((.....))))((((.((((((((((........))))..............)))))))))).......... ( -8.67 = -9.27 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:41 2006