
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,496,959 – 6,497,072 |
| Length | 113 |
| Max. P | 0.806640 |

| Location | 6,496,959 – 6,497,049 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -23.02 |
| Energy contribution | -24.33 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
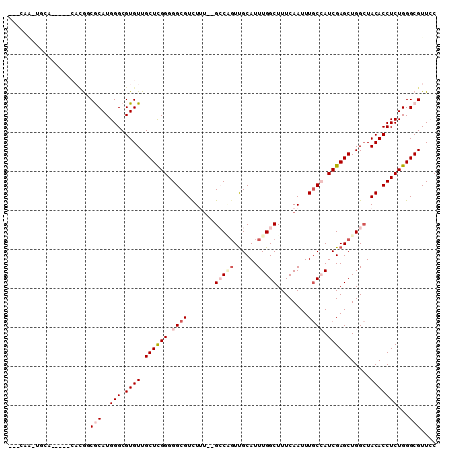
>3R_DroMel_CAF1 6496959 90 + 27905053 ------------------CAUCGCAUGGGCGUGUUGCUCGGGGGCGUCUUU--GCCAGUUGCAUUUGGCUUUCAAUUUGCCAUCGAGCUGGGUACACCUCUGGGCGUUCC ------------------...(((..(((.((((.(((((..((((.....--(((((......)))))........))))..))))).....)))))))...))).... ( -32.82) >DroVir_CAF1 298159 106 + 1 --CCAGCAGCAAUAUUCAAAGCACAUGGGCGUGUUGCUCGGGGGCGUCUGU--GGCUCUCGCAUUUGGCUUUCAAUUUACAAUCGAGCUGGCUACACCUCUGGGCGUUGC --...(((((.........(((((......)))))(((((((((....(((--((((((((...(((............))).))))..))))))))))))))))))))) ( -37.70) >DroPse_CAF1 268334 106 + 1 --GCAAUUGCAUGCCACAUGCCGCAUGGGCGUGUUGCUUGGGGGCGUCUUU--GCCGGUUGCAUUUGGCUUUCAAUUUGCCAUCGAGCUGGCUACACCUCUGGGCGUUCC --......(((((((.((((...))))))))))).(((..((((.((....--(((((((.(...((((.........))))..)))))))).)).))))..)))..... ( -43.90) >DroEre_CAF1 273631 90 + 1 ------------------CGGCGCAUGGGCGUGUUGCUCGGGGGCGUCUUA--GCCAGUUGCAUUUGGCUUUCAAUUUGCCAUCGAGCUGGCUACACCUCUGGGCGUUCC ------------------..((((....))))...(((((((((.((...(--(((((((.(...((((.........))))..))))))))))).)))))))))..... ( -39.00) >DroWil_CAF1 380182 110 + 1 CAUCAACUUCAACAUCCACGGCACAUGGGCGUGUUGCUUGGGGGCGUUUGUGUGGCUGUUGCAUUUGGCUUUCAAUUUGCAAUCGAGCUGGCUACACCUCUGGGCGUUGC .........((((...((((.(.....).))))..(((..((((.((..((.(((((((((((.(((.....)))..))))))..))))))).)).))))..))))))). ( -35.30) >DroPer_CAF1 273049 106 + 1 --GCAAUUGCAUGCCACAUGCCGCAUGGGCGUGUUGCUUGGGGGCGUCUUU--GCCGGUUGCAUUUGGCUUUCAAUUUGCCAUCGAGCUGGCUACACCUCUGGGCGUUCC --......(((((((.((((...))))))))))).(((..((((.((....--(((((((.(...((((.........))))..)))))))).)).))))..)))..... ( -43.90) >consensus ___CAA_UGCA_____CACGGCGCAUGGGCGUGUUGCUCGGGGGCGUCUUU__GCCAGUUGCAUUUGGCUUUCAAUUUGCCAUCGAGCUGGCUACACCUCUGGGCGUUCC .....................(((..(((.((((.((((((.((((.......(((((......)))))........)))).)))))).....)))))))...))).... (-23.02 = -24.33 + 1.31)

| Location | 6,496,973 – 6,497,072 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.88 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -23.84 |
| Energy contribution | -25.20 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
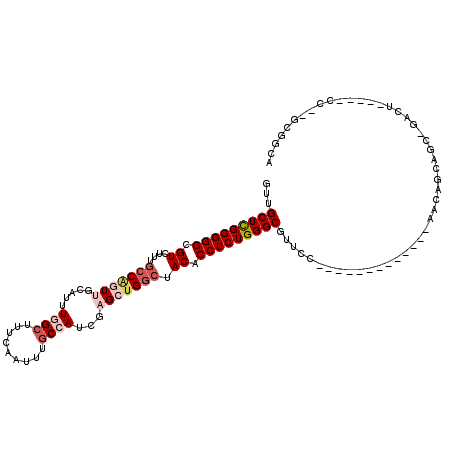
>3R_DroMel_CAF1 6496973 99 + 27905053 GUUGCUCGGGGGCGUCUUUGCCAGUUGCAUUUGGCUUUCAAUUUGCCAUCGAGCUGGGUACACCUCUGGGCGUUCC-------------AAGAUCCGU-GACU-----CCGAGAGGCA ...(((((..((((.....(((((......)))))........))))..)))))........(((((.((.((..(-------------.......).-.)).-----)).))))).. ( -35.52) >DroVir_CAF1 298189 85 + 1 GUUGCUCGGGGGCGUCUGUGGCUCUCGCAUUUGGCUUUCAAUUUACAAUCGAGCUGGCUACACCUCUGGGCGUUGC-------------AGCAGCAAC-------------------- ...(((((((((....(((((((((((...(((............))).))))..))))))))))))))))(((((-------------....)))))-------------------- ( -32.30) >DroPse_CAF1 268364 89 + 1 GUUGCUUGGGGGCGUCUUUGCCGGUUGCAUUUGGCUUUCAAUUUGCCAUCGAGCUGGCUACACCUCUGGGCGUUCC-------------AGCAGCA----------------GCAGCA ((((((..(((((((((..(((((((.(...((((.........))))..)))))))).........)))))))))-------------...))))----------------)).... ( -35.90) >DroEre_CAF1 273645 99 + 1 GUUGCUCGGGGGCGUCUUAGCCAGUUGCAUUUGGCUUUCAAUUUGCCAUCGAGCUGGCUACACCUCUGGGCGUUCC-------------GAGAUCCGC-GACU-----CCAAGGGGCA ...(((((((((.((...((((((((.(...((((.........))))..))))))))))).)))))))))(((((-------------(((......-..))-----)...))))). ( -40.90) >DroYak_CAF1 269279 112 + 1 GUUGCUCGGGGGCGUCUUUGCCAGUUGCAUUUGGCUUUCAAUUUGCCAUCGAGCUGGCUACACCUCUGGGCGUUUCUCCGGCGUUCCUGCAGAUCUGC-GACU-----CCAAGGGGCA ...(((((((((.((....(((((((.(...((((.........))))..)))))))).)).)))))))))((..((..((.(((...(((....)))-))).-----)).))..)). ( -44.90) >DroMoj_CAF1 361384 102 + 1 GUUGCUCGGGGGCGUCUGGGCCGCUCACAUUUGGCUUUCAAUUUGCAAUUGCGCUGGCUACACCUCUGGGCGUUGC-------------AACAGCAUCCGACUUAUCUGC---CAGCA (((((..((((((......))).)))....(((.....)))...)))))...((((((........(((..((((.-------------..))))..)))........))---)))). ( -28.99) >consensus GUUGCUCGGGGGCGUCUUUGCCAGUUGCAUUUGGCUUUCAAUUUGCCAUCGAGCUGGCUACACCUCUGGGCGUUCC_____________AACAGCAGC_GACU_____CC__GCGGCA ...(((((((((.((....(((((((.....((((.........))))...))))))).)).)))))))))............................................... (-23.84 = -25.20 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:36 2006