
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,428,095 – 6,428,253 |
| Length | 158 |
| Max. P | 0.999776 |

| Location | 6,428,095 – 6,428,203 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -43.32 |
| Consensus MFE | -32.56 |
| Energy contribution | -32.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6428095 108 + 27905053 GAAGUCACAUCGCAUAUUUGUGACAUAUGGCAGCAAGACAUUGUUGAUCUUGCUGCGGC-AUGGGA-----------AGAUGCUGCUGCGGCAACGUAGAUGCUGCUCUUGUCAGCGGGG ...((((((.........)))))).....(((((((((((....)).))))))))).((-.(((.(-----------(((.((..(((((....))))).....)))))).))))).... ( -43.70) >DroSec_CAF1 191745 108 + 1 GAAGUCACAUCGCAUAUUUGUGACAUAUGGCAGCAAGACAUUGUUGAUCUUGCUGCGGC-AUGGGA-----------AGAAGCUGCUGCGGCAACGUGGAUGCUGCUCUUGUCAGCGGGG ...((((((.........)))))).....(((((((((((....)).))))))))).((-.(((.(-----------((((((..(..((....))..)..)))..)))).))))).... ( -43.00) >DroSim_CAF1 216977 108 + 1 GAAGUCACAUCGCAUAUUUGUGACAUAUGGCAGCAAGACAUUGUUGAUCUUGCUGCGGC-AUGGGA-----------AGAAGCUGCUGCGGCAACGUGGAUGCUGCUCUUGUCAGCGGGG ...((((((.........)))))).....(((((((((((....)).))))))))).((-.(((.(-----------((((((..(..((....))..)..)))..)))).))))).... ( -43.00) >DroEre_CAF1 203139 108 + 1 GAAGUCACAUCGCAUAUUUGUGACAUAUGGCAGCAAGACAUUGUUGAUCUGGCUGCGGCAAUGGGA-----------AGAUGUGGCUGCAGCAUCGUAGAUGUUGCUCUUGUCAGCGG-G ..((((((((((((....)))..(((.(.(((((.(((((....)).))).))))).)..)))...-----------.)))))))))((((((((...)))))))).((((....)))-) ( -37.60) >DroYak_CAF1 196616 118 + 1 GAAGUCACAUCGCAUAUUUGUGACAUAUGGCAGCAAGACAUUGUUGAUCUUGCUGCGGG-AUGGGAUGUGGGAUUUGGGAUGUGG-UGCCGCAUCGUAGAUGUUGCUCUUGUCAGCGGGG ...((((((.........)))))).....(((((((((((....)).)))))))))..(-(..(((.(..(.(((((.((((((.-...)))))).))))).)..))))..))....... ( -49.30) >consensus GAAGUCACAUCGCAUAUUUGUGACAUAUGGCAGCAAGACAUUGUUGAUCUUGCUGCGGC_AUGGGA___________AGAUGCUGCUGCGGCAACGUAGAUGCUGCUCUUGUCAGCGGGG ...((((((.........)))))).....(((((((((((....)).))))))))).........................(((((.((((((.......))))))....).)))).... (-32.56 = -32.96 + 0.40)

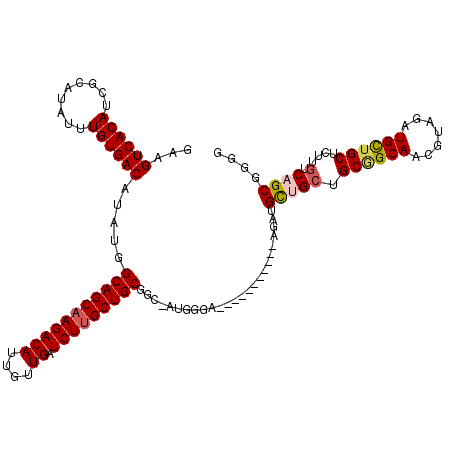
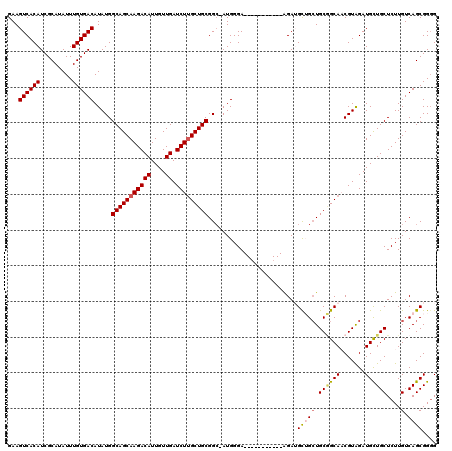
| Location | 6,428,095 – 6,428,203 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -28.50 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6428095 108 - 27905053 CCCCGCUGACAAGAGCAGCAUCUACGUUGCCGCAGCAGCAUCU-----------UCCCAU-GCCGCAGCAAGAUCAACAAUGUCUUGCUGCCAUAUGUCACAAAUAUGCGAUGUGACUUC ....((((....(.(((((......)))))).)))).((((..-----------....))-)).((((((((((.......)))))))))).....((((((.........))))))... ( -36.20) >DroSec_CAF1 191745 108 - 1 CCCCGCUGACAAGAGCAGCAUCCACGUUGCCGCAGCAGCUUCU-----------UCCCAU-GCCGCAGCAAGAUCAACAAUGUCUUGCUGCCAUAUGUCACAAAUAUGCGAUGUGACUUC ....((((....(.(((((......))))))....))))....-----------......-...((((((((((.......)))))))))).....((((((.........))))))... ( -34.40) >DroSim_CAF1 216977 108 - 1 CCCCGCUGACAAGAGCAGCAUCCACGUUGCCGCAGCAGCUUCU-----------UCCCAU-GCCGCAGCAAGAUCAACAAUGUCUUGCUGCCAUAUGUCACAAAUAUGCGAUGUGACUUC ....((((....(.(((((......))))))....))))....-----------......-...((((((((((.......)))))))))).....((((((.........))))))... ( -34.40) >DroEre_CAF1 203139 108 - 1 C-CCGCUGACAAGAGCAACAUCUACGAUGCUGCAGCCACAUCU-----------UCCCAUUGCCGCAGCCAGAUCAACAAUGUCUUGCUGCCAUAUGUCACAAAUAUGCGAUGUGACUUC .-..((((.....((((.(......).)))).))))((((((.-----------..........(((((.((((.......)))).)))))(((((.......))))).))))))..... ( -28.70) >DroYak_CAF1 196616 118 - 1 CCCCGCUGACAAGAGCAACAUCUACGAUGCGGCA-CCACAUCCCAAAUCCCACAUCCCAU-CCCGCAGCAAGAUCAACAAUGUCUUGCUGCCAUAUGUCACAAAUAUGCGAUGUGACUUC ..((((.....(((......))).....))))..-...............((((((....-...((((((((((.......))))))))))(((((.......))))).))))))..... ( -32.20) >consensus CCCCGCUGACAAGAGCAGCAUCUACGUUGCCGCAGCAGCAUCU___________UCCCAU_GCCGCAGCAAGAUCAACAAUGUCUUGCUGCCAUAUGUCACAAAUAUGCGAUGUGACUUC ....((((....(.(((((......)))))).))))............................((((((((((.......)))))))))).....((((((.........))))))... (-28.50 = -29.38 + 0.88)

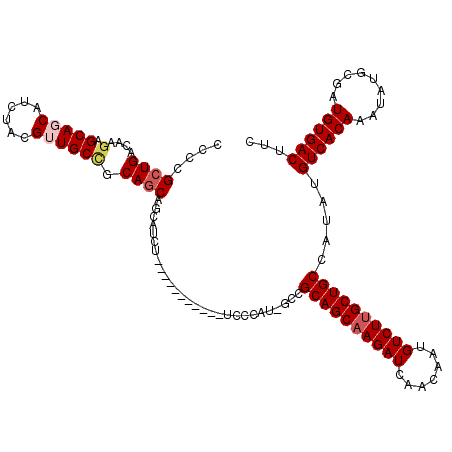
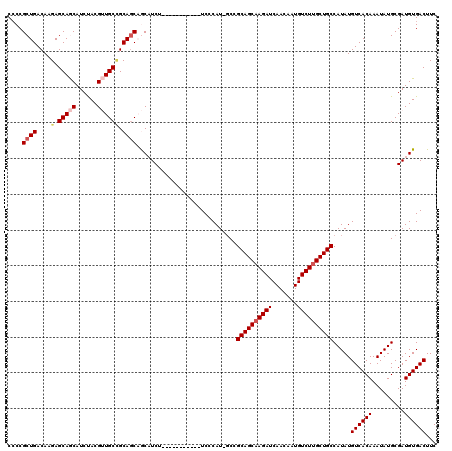
| Location | 6,428,135 – 6,428,243 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -31.59 |
| Energy contribution | -32.87 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6428135 108 + 27905053 UUGUUGAUCUUGCUGCGGC-AUGGGA-----------AGAUGCUGCUGCGGCAACGUAGAUGCUGCUCUUGUCAGCGGGGACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGAAUGU ..((((....(((.(((((-((....-----------..))))))).))).))))((((...))))(((((..(((((((.(((((......)))))....)))))))...))))).... ( -39.70) >DroSec_CAF1 191785 107 + 1 UUGUUGAUCUUGCUGCGGC-AUGGGA-----------AGAAGCUGCUGCGGCAACGUGGAUGCUGCUCUUGUCAGCGGGGACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGAAUG- ..(((.((.((((((((((-(.(...-----------.....)))))))))))).)).)))....(((..((.(((((((.(((((......)))))....))))))).)).)))....- ( -41.30) >DroSim_CAF1 217017 107 + 1 UUGUUGAUCUUGCUGCGGC-AUGGGA-----------AGAAGCUGCUGCGGCAACGUGGAUGCUGCUCUUGUCAGCGGGGACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGAAUG- ..(((.((.((((((((((-(.(...-----------.....)))))))))))).)).)))....(((..((.(((((((.(((((......)))))....))))))).)).)))....- ( -41.30) >DroEre_CAF1 203179 107 + 1 UUGUUGAUCUGGCUGCGGCAAUGGGA-----------AGAUGUGGCUGCAGCAUCGUAGAUGUUGCUCUUGUCAGCGG-GACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGACUG- ......(((((((((((((.(((...-----------...))).))))))))....)))))....(((..((.(((((-(.(((((......))))).....)))))).)).)))....- ( -33.10) >DroYak_CAF1 196656 117 + 1 UUGUUGAUCUUGCUGCGGG-AUGGGAUGUGGGAUUUGGGAUGUGG-UGCCGCAUCGUAGAUGUUGCUCUUGUCAGCGGGGACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGACCG- ...............((((-(..(((.(..(.(((((.((((((.-...)))))).))))).)..))))..))(((((((.(((((......)))))....)))))))........)))- ( -46.70) >consensus UUGUUGAUCUUGCUGCGGC_AUGGGA___________AGAUGCUGCUGCGGCAACGUAGAUGCUGCUCUUGUCAGCGGGGACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGAAUG_ ..(((.((.((((((((((.........................)))))))))).)).)))....(((..((.(((((((.(((((......)))))....))))))).)).)))..... (-31.59 = -32.87 + 1.28)


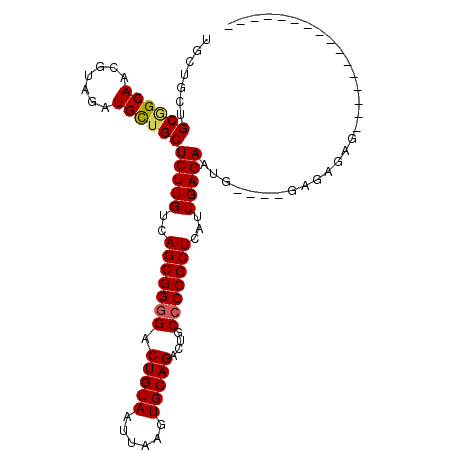
| Location | 6,428,163 – 6,428,253 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.44 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6428163 90 + 27905053 UGCUGCUGCGGCAACGUAGAUGCUGCUCUUGUCAGCGGGGACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGAAUGUAGAGAGAGAG---------------- .....(((((....)))))...(((((((((..(((((((.(((((......)))))....)))))))...)))))..))))........---------------- ( -32.90) >DroSec_CAF1 191813 86 + 1 AGCUGCUGCGGCAACGUGGAUGCUGCUCUUGUCAGCGGGGACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGAAUG----GAGAGAG---------------- (((..(..((....))..)..))).(((((...(((((((.(((((......)))))....)))))))(((.....)))----)))))..---------------- ( -33.00) >DroSim_CAF1 217045 86 + 1 AGCUGCUGCGGCAACGUGGAUGCUGCUCUUGUCAGCGGGGACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGAAUG----GAGAGAG---------------- (((..(..((....))..)..))).(((((...(((((((.(((((......)))))....)))))))(((.....)))----)))))..---------------- ( -33.00) >DroEre_CAF1 203208 101 + 1 UGUGGCUGCAGCAUCGUAGAUGUUGCUCUUGUCAGCGG-GACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGACUG----GAGAUCGGUAUGGUAUGGUAUUG ..((((.((((((((...))))))))....))))((((-(.(((((......))))).....)))))((((....((((----.....)))).....))))..... ( -29.30) >DroYak_CAF1 196695 101 + 1 UGUGG-UGCCGCAUCGUAGAUGUUGCUCUUGUCAGCGGGGACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGACCG----GAGAGCGGAAUCGGAUGGUAUGG .(((.-...)))(((((.(((.((((((((((((((((((.(((((......)))))....)))))))......)))..----)))))))).)))..))))).... ( -37.50) >consensus UGCUGCUGCGGCAACGUAGAUGCUGCUCUUGUCAGCGGGGACUGUAAUUAAGUGCAGACUGCCCCGCUCAUUGAGAAUG____GAGAGAG________________ .......((((((.......))))))(((((..(((((((.(((((......)))))....)))))))...))))).............................. (-26.40 = -26.44 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:16 2006