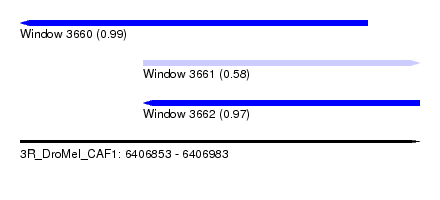
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,406,853 – 6,406,983 |
| Length | 130 |
| Max. P | 0.986226 |

| Location | 6,406,853 – 6,406,966 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.38 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6406853 113 - 27905053 UAAUAUCUUUCUUUAGGUGUUCAGUAAAA-----UCGGAUUUAUCGAUUAUAGAUUUUCGAAAUUACCUUAAGAGAGCUUUACUACAUUGCUCUCAAUGAUCUUGUAACUUCUAUAAA .............((((.(((((((((..-----(((((...(((.......))).)))))..)))).....((((((...........))))))........)).))).)))).... ( -20.80) >DroPse_CAF1 169259 94 - 1 -----ACUCUCUUUAAGUUUUCAUGGAAAUCAAAAC----------------GAUUUCUGGAAUAAUUUCGGGAGAGUUACGAUUAUUUGCAUGGGUUGAACCUGGAACUA---AAAC -----(((((((..((((((((.((((((((.....----------------))))))))))).)))))..))))))).....(((.((.((.((......)))).)).))---)... ( -23.80) >DroSec_CAF1 170630 117 - 1 U-AUAUCUUUCUUUAGGGGUUUAGGAAAAUCAAAUCGGAAUUUUCGAUUAUAGAUUUUCGAAACUACCUUAAGAGAGCUUUACUACAUUGCUCUCAACGAUCUUGGAACUUUUAUAAA (-(((...((((((((((((((..(((((((.(((((((...)))))))...))))))).))))..))))))((((((...........)))))).........))))....)))).. ( -27.80) >DroSim_CAF1 194780 117 - 1 U-AUAUCUUUCUUUAGGUGGUUAUGAAAAUCGAAUCGGAAUUUUCGAUUAUAGAUUUUCGAAAUGACCUUAAGGGAGCUUUACUACAUUGCUCUCAACGAUCUUGGAACUUUUAUAAA (-(((...((((..((((.(((.((((((((.(((((((...)))))))...)))))))).))).))))...((((((...........)))))).........))))....)))).. ( -30.60) >DroEre_CAF1 181854 117 - 1 U-AUUUCCGUCUUUUGGUGUUCAGGAAAAUCGGAUCGGAAUUUUCGAUUAUCGGUUUUUGAAAUGACCUCAAGAGAGCUGUACUCACUUGCUGCCAUUGCUCAUGAACCUUUUAUAAA .-....(((.....))).......((.(((((((........))))))).))(((((.(((((((.(..((((.(((.....))).))))..).))))..))).)))))......... ( -23.80) >DroYak_CAF1 174752 117 - 1 U-AUUUCCACCUUUAGGUCUUCUGGAAAAUCGGAUCGGAAUUUUCGAUUAUCGGUUUUUGAAAUGACCUCAAGGGAGUUUUACUAAGCUGCUCGCUUUGAUCCUGGAACAUUUAUUAA .-..((((.((((.(((((...(.(((((((((((((((...)))))))..)))))))).)...))))).)))).((..(((..((((.....)))))))..)))))).......... ( -32.10) >consensus U_AUAUCUUUCUUUAGGUGUUCAGGAAAAUCGAAUCGGAAUUUUCGAUUAUAGAUUUUCGAAAUGACCUCAAGAGAGCUUUACUAAAUUGCUCUCAAUGAUCUUGGAACUUUUAUAAA ......(((((((.(((((.....(((((((.((((((.....))))))...))))))).....))))).)))))))......................................... (-15.80 = -16.58 + 0.79)



| Location | 6,406,893 – 6,406,983 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -16.30 |
| Consensus MFE | -6.82 |
| Energy contribution | -6.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6406893 90 + 27905053 GCUCUCUUAAGGUAAUUUCGAAAAUCUAUAAUCGAUAAAUCCGA-----UUUUACUGAACACCUAAAGAAAGAUAUUACUACACUACCAUCCUAA .((.((((.((((..((...((((((................))-----))))...))..)))).)))).))....................... ( -10.49) >DroSec_CAF1 170670 92 + 1 GCUCUCUUAAGGUAGUUUCGAAAAUCUAUAAUCGAAAAUUCCGAUUUGAUUUUCCUAAACCCCUAAAGAAAGAUAU-AAUGCACUACCAUCU--G .((.((((.(((..((((.(((((((...(((((.......))))).)))))))..)))).))).)))).))....-...............--. ( -18.30) >DroSim_CAF1 194820 94 + 1 GCUCCCUUAAGGUCAUUUCGAAAAUCUAUAAUCGAAAAUUCCGAUUCGAUUUUCAUAACCACCUAAAGAAAGAUAU-ACUGCACUACUAUCCUAA .((..(((.((((..((..(((((((...(((((.......))))).)))))))..))..)))).)))..))....-.................. ( -15.40) >DroEre_CAF1 181894 94 + 1 GCUCUCUUGAGGUCAUUUCAAAAACCGAUAAUCGAAAAUUCCGAUCCGAUUUUCCUGAACACCAAAAGACGGAAAU-ACUGCACUAGCAGCCAAA .((.((((..(((...((((((((.((...((((.......)))).)).))))..)))).)))..)))).))....-.((((....))))..... ( -15.60) >DroYak_CAF1 174792 94 + 1 ACUCCCUUGAGGUCAUUUCAAAAACCGAUAAUCGAAAAUUCCGAUCCGAUUUUCCAGAAGACCUAAAGGUGGAAAU-ACUGCACUAGUUUCCAAA ....((((.(((((.(((..((((.((...((((.......)))).)).))))..))).))))).))))(((((((-.........))))))).. ( -21.70) >consensus GCUCUCUUAAGGUCAUUUCGAAAAUCUAUAAUCGAAAAUUCCGAUCCGAUUUUCCUGAACACCUAAAGAAAGAUAU_ACUGCACUACCAUCCAAA .((.((((.((((..((...((((......((((.......))))....))))...))..)))).)))).))....................... ( -6.82 = -6.90 + 0.08)


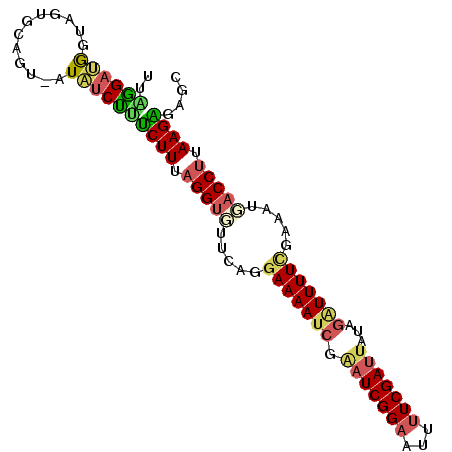
| Location | 6,406,893 – 6,406,983 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -16.18 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6406893 90 - 27905053 UUAGGAUGGUAGUGUAGUAAUAUCUUUCUUUAGGUGUUCAGUAAAA-----UCGGAUUUAUCGAUUAUAGAUUUUCGAAAUUACCUUAAGAGAGC .......................(((((((.(((((....(.((((-----((.((((....))))...))))))).....))))).))))))). ( -16.70) >DroSec_CAF1 170670 92 - 1 C--AGAUGGUAGUGCAUU-AUAUCUUUCUUUAGGGGUUUAGGAAAAUCAAAUCGGAAUUUUCGAUUAUAGAUUUUCGAAACUACCUUAAGAGAGC .--.((((......))))-....(((((((.((((((((..(((((((.(((((((...)))))))...))))))).))))).))).))))))). ( -25.30) >DroSim_CAF1 194820 94 - 1 UUAGGAUAGUAGUGCAGU-AUAUCUUUCUUUAGGUGGUUAUGAAAAUCGAAUCGGAAUUUUCGAUUAUAGAUUUUCGAAAUGACCUUAAGGGAGC ..................-....((..(((.((((.(((.((((((((.(((((((...)))))))...)))))))).))).)))).)))..)). ( -28.10) >DroEre_CAF1 181894 94 - 1 UUUGGCUGCUAGUGCAGU-AUUUCCGUCUUUUGGUGUUCAGGAAAAUCGGAUCGGAAUUUUCGAUUAUCGGUUUUUGAAAUGACCUCAAGAGAGC ....(((((....)))))-.......((((((((.((.((.(((((((((((((((...)))))))..))))))))....)))).)))))))).. ( -26.50) >DroYak_CAF1 174792 94 - 1 UUUGGAAACUAGUGCAGU-AUUUCCACCUUUAGGUCUUCUGGAAAAUCGGAUCGGAAUUUUCGAUUAUCGGUUUUUGAAAUGACCUCAAGGGAGU ..((((((((.....)))-..)))))((((.(((((...(.(((((((((((((((...)))))))..)))))))).)...))))).)))).... ( -28.70) >consensus UUAGGAUGGUAGUGCAGU_AUAUCUUUCUUUAGGUGUUCAGGAAAAUCGAAUCGGAAUUUUCGAUUAUAGAUUUUCGAAAUGACCUUAAGAGAGC ..((((((............))))))((((.(((((.....(((((((.(((((((...)))))))...))))))).....))))).)))).... (-16.18 = -16.50 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:04 2006