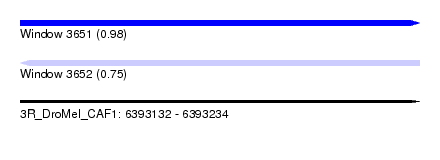
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,393,132 – 6,393,234 |
| Length | 102 |
| Max. P | 0.979601 |

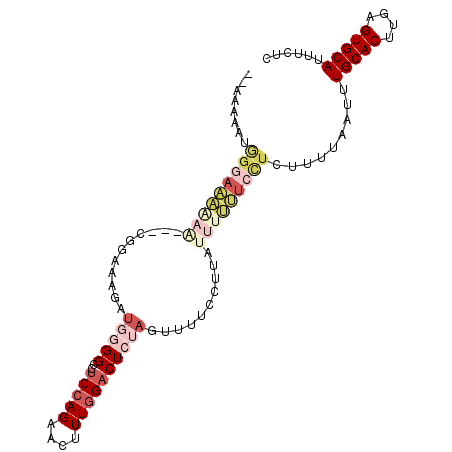
| Location | 6,393,132 – 6,393,234 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -11.81 |
| Energy contribution | -14.32 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6393132 102 + 27905053 AAAAAAAUGGGAAAAAAUGUCGGAGAGAUGGGGUCUCCAGAACUUUGGACUCUAGUUUUCCUUAUUUUUUCCUCUUUUAAUUUGCACCUGAGUGCAUUUCUC ...((((.(((((((((((..((((((.((((((((..........)))))))).)))))).))))))))))).))))....(((((....)))))...... ( -32.30) >DroPse_CAF1 155961 94 + 1 --AGAGA-GGGAGGGAG---GGGUACGGUAGGGAAGCAAGAACUUUGGACUGCAUUUU--GUAGUUUCCUCUUUUUUUAAUUUGCACUUGAGUGCAUUUCUC --.((((-(((((((((---(((.........((((......))))(((((((.....--))))))))))))))))))....(((((....))))).))))) ( -26.40) >DroSec_CAF1 157014 100 + 1 --AAAAAUGGGAAAAAAUGUCGGAGAGAUGGGGCCUCCAGAACUUUGGACUCUAGUUUUCCUUAUUUUUUCCUCUUUUAAUUUGCACUUGAGUGCAUUUCUC --.((((.(((((((((((..((((((.(((((..(((((....)))))))))).)))))).))))))))))).))))....(((((....)))))...... ( -32.60) >DroSim_CAF1 180453 102 + 1 AAAAAAAUGGGAAAAAAUGUCGGAGAGAUGGGGCCUCCAGAACUUUGGACUCUAGUUUUCCUUAUUUUUUCCUCUUUUAAUUUGCACUUGAGUGCAUUUCUC ...((((.(((((((((((..((((((.(((((..(((((....)))))))))).)))))).))))))))))).))))....(((((....)))))...... ( -32.60) >DroEre_CAF1 168141 79 + 1 -----------------------AAAGAUGGGGCCUCCAGAACUUUGGACUCUAGUUCUCUUGAUUUUUUCCUCUUUUAAUUUGCACUCGAGUGCAUUUCUC -----------------------(((((.((((..((.((((((.........))))))...))....))))))))).....(((((....)))))...... ( -14.40) >DroYak_CAF1 160461 88 + 1 -----AACAGCAAA---------AAAAAACAGGCCUCCAGAACUUUGGACUCUAGUUUUCCUUAUUUUUUCCUCUUUUAAUUUGCACUUGAGUGCAUUUCUC -----.........---------..(((((.((..(((((....)))))..)).))))).......................(((((....)))))...... ( -13.50) >consensus __AAAAAUGGGAAAAAA___CGGAAAGAUGGGGCCUCCAGAACUUUGGACUCUAGUUUUCCUUAUUUUUUCCUCUUUUAAUUUGCACUUGAGUGCAUUUCUC ........(((((((((...........(((((..(((((....))))))))))..........))))))))).........(((((....)))))...... (-11.81 = -14.32 + 2.50)

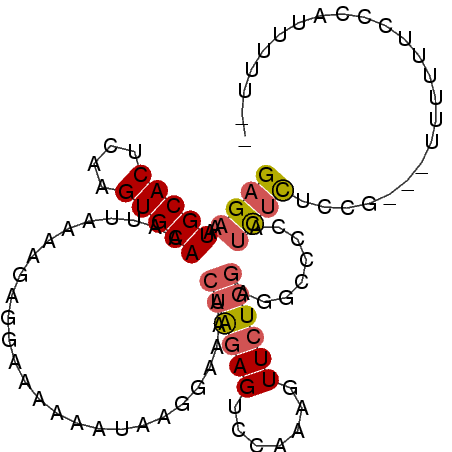
| Location | 6,393,132 – 6,393,234 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -20.02 |
| Consensus MFE | -10.14 |
| Energy contribution | -10.45 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6393132 102 - 27905053 GAGAAAUGCACUCAGGUGCAAAUUAAAAGAGGAAAAAAUAAGGAAAACUAGAGUCCAAAGUUCUGGAGACCCCAUCUCUCCGACAUUUUUUCCCAUUUUUUU (((((((((((....)))).........(.(((((((((..(((.........)))...(((..(((((.......))))))))))))))))))))))))). ( -23.90) >DroPse_CAF1 155961 94 - 1 GAGAAAUGCACUCAAGUGCAAAUUAAAAAAAGAGGAAACUAC--AAAAUGCAGUCCAAAGUUCUUGCUUCCCUACCGUACCC---CUCCCUCCC-UCUCU-- ((((..(((((....)))))...........((((..(((.(--.....).)))...((((....))))............)---)))......-)))).-- ( -15.90) >DroSec_CAF1 157014 100 - 1 GAGAAAUGCACUCAAGUGCAAAUUAAAAGAGGAAAAAAUAAGGAAAACUAGAGUCCAAAGUUCUGGAGGCCCCAUCUCUCCGACAUUUUUUCCCAUUUUU-- ......(((((....)))))....((((..(((((((((..(((...((((((.......)))))).(....).....)))...)))))))))..)))).-- ( -22.70) >DroSim_CAF1 180453 102 - 1 GAGAAAUGCACUCAAGUGCAAAUUAAAAGAGGAAAAAAUAAGGAAAACUAGAGUCCAAAGUUCUGGAGGCCCCAUCUCUCCGACAUUUUUUCCCAUUUUUUU (((((((((((....)))).........(.(((((((((..(((...((((((.......)))))).(....).....)))...))))))))))))))))). ( -22.90) >DroEre_CAF1 168141 79 - 1 GAGAAAUGCACUCGAGUGCAAAUUAAAAGAGGAAAAAAUCAAGAGAACUAGAGUCCAAAGUUCUGGAGGCCCCAUCUUU----------------------- ......(((((....))))).....(((((((......((...((((((.........)))))).))....)).)))))----------------------- ( -15.00) >DroYak_CAF1 160461 88 - 1 GAGAAAUGCACUCAAGUGCAAAUUAAAAGAGGAAAAAAUAAGGAAAACUAGAGUCCAAAGUUCUGGAGGCCUGUUUUUU---------UUUGCUGUU----- ......(((((....))))).......(((((((((((((.((....((((((.......))))))...))))))))))---------))).))...----- ( -19.70) >consensus GAGAAAUGCACUCAAGUGCAAAUUAAAAGAGGAAAAAAUAAGGAAAACUAGAGUCCAAAGUUCUGGAGGCCCCAUCUCUCCG___UUUUUUCCCAUUUUU__ ((((..(((((....)))))...........................((((((.......))))))........))))........................ (-10.14 = -10.45 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:55 2006