
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,388,026 – 6,388,128 |
| Length | 102 |
| Max. P | 0.803005 |

| Location | 6,388,026 – 6,388,128 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.78 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.98 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6388026 102 + 27905053 GA----CUGUCUGCACGACUGUCAAGUUGUCAAAAGUGCCACGGCCUGGCCAUAUUGUGAACAAACAAAUGUUCAAG--AUUGUCGUGGCCUAGGCCUAGAGCAAA---AU------ .(----((...((.((((((....))))))))..)))(((..((((((((((((((.((((((......)))))).)--))....)))))).)))))..).))...---..------ ( -35.30) >DroVir_CAF1 163231 106 + 1 GAGUCUUAGUCUGCACGACUGUCAAGUUGUCAAAACUGCCAUGGCCCGGCCAUAUUAUGGCCAAACAAAUGCCCCAACAACGAUCGUGGCCUAGGCCUAGACAACA----------- ..((((..(((((.((((((....)))))).......((((((..(.((((((...)))))).......((......))..)..)))))).)))))..))))....----------- ( -32.90) >DroGri_CAF1 193765 114 + 1 GAGUCUUGGUCUGCACGACUGUCAAGUUGUCAAAAGUGCCAUGGCCCGGCCAUAUUAUGGCUAAACAAAUGCCCCA---ACGAUCGUGGCCUAGGCCUAGACGAUCAAAACAACCAU ..((((.((((((.((((((....)))))))...((.((((((..(.((((((...))))))........(.....---.))..))))))))))))).))))............... ( -33.50) >DroMoj_CAF1 211317 106 + 1 GGGUCCGUGUCUGCACGACUGUCAAGUUGUCAAAACUGCCAUGGCUUGGCCAUAUUCUGGCCUAACAAAUGCCCAAACAGCGAUCGUGGCCUAGGCCUAGACCGGC----------- ....(((.(((((.((((((....))))))............(((((((((((..((((......))...((.......))))..)))))).))))))))))))).----------- ( -33.50) >DroAna_CAF1 150969 102 + 1 GA----CUGUCUGCACGACUGUCAAGUUGUCAAAAGUGCCACGGCCCGGCCAUAUUGUGGGCAAACAAAUGUUCGAG--AUUGUCGUGGCCUAGGCCUAGAACGAA---AC------ ((----(.(((.....))).)))...((((....((.(((..((((((((...(((.((((((......)))))).)--)).)))).))))..)))))...)))).---..------ ( -30.80) >DroPer_CAF1 157812 101 + 1 GA----CUGUCUGCACGACUGUCAAGUUGUCAAAACUGCCACGGCCUGGCCAUAUUGUGGCCAAACAAAUGCUCAAG--AUUGUCGUGGCCUAGGCCUAGACCAGA---G------- ..----(((((((.(((((.(((.((((.....))))((....(..(((((((...)))))))..)....))....)--)).)))))(((....))))))).))).---.------- ( -32.70) >consensus GA____CUGUCUGCACGACUGUCAAGUUGUCAAAACUGCCACGGCCCGGCCAUAUUGUGGCCAAACAAAUGCCCAAG__ACGAUCGUGGCCUAGGCCUAGACCAAA___A_______ ........(((((.((((((....)))))).......(((..((((.((((((...)))))).................((....))))))..))).)))))............... (-22.82 = -22.98 + 0.17)

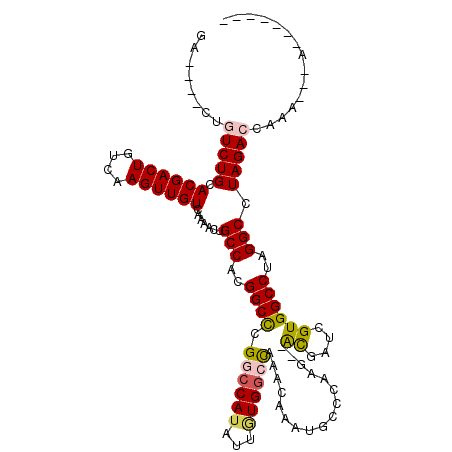

| Location | 6,388,026 – 6,388,128 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.78 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6388026 102 - 27905053 ------AU---UUUGCUCUAGGCCUAGGCCACGACAAU--CUUGAACAUUUGUUUGUUCACAAUAUGGCCAGGCCGUGGCACUUUUGACAACUUGACAGUCGUGCAGACAG----UC ------..---..((((...(((((.(((((.(.....--).((((((......)))))).....))))))))))..)))).............(((.(((.....))).)----)) ( -32.40) >DroVir_CAF1 163231 106 - 1 -----------UGUUGUCUAGGCCUAGGCCACGAUCGUUGUUGGGGCAUUUGUUUGGCCAUAAUAUGGCCGGGCCAUGGCAGUUUUGACAACUUGACAGUCGUGCAGACUAAGACUC -----------.((((((..(((....)))......(((((..((((......((((((((...))))))))((....)).))))..)))))..)))((((.....))))..))).. ( -37.60) >DroGri_CAF1 193765 114 - 1 AUGGUUGUUUUGAUCGUCUAGGCCUAGGCCACGAUCGU---UGGGGCAUUUGUUUAGCCAUAAUAUGGCCGGGCCAUGGCACUUUUGACAACUUGACAGUCGUGCAGACCAAGACUC ......((((((...((((.(((((.(((((.....((---((.(((.........))).)))).))))))))))...((((..(((.(.....).)))..)))))))))))))).. ( -36.50) >DroMoj_CAF1 211317 106 - 1 -----------GCCGGUCUAGGCCUAGGCCACGAUCGCUGUUUGGGCAUUUGUUAGGCCAGAAUAUGGCCAAGCCAUGGCAGUUUUGACAACUUGACAGUCGUGCAGACACGGACCC -----------...((((..(((....))).....((.((((((.((..(((((((((((.....)))))..((....)).............))))))..)).)))))))))))). ( -39.40) >DroAna_CAF1 150969 102 - 1 ------GU---UUCGUUCUAGGCCUAGGCCACGACAAU--CUCGAACAUUUGUUUGCCCACAAUAUGGCCGGGCCGUGGCACUUUUGACAACUUGACAGUCGUGCAGACAG----UC ------((---((.......(((((.(((((.......--..(((((....))))).........))))))))))...((((..(((.(.....).)))..))))))))..----.. ( -29.47) >DroPer_CAF1 157812 101 - 1 -------C---UCUGGUCUAGGCCUAGGCCACGACAAU--CUUGAGCAUUUGUUUGGCCACAAUAUGGCCAGGCCGUGGCAGUUUUGACAACUUGACAGUCGUGCAGACAG----UC -------.---....((((.(((....)))(((((...--.....((....(((((((((.....)))))))))....)).(((..(....)..))).)))))..))))..----.. ( -34.20) >consensus _______U___UCUGGUCUAGGCCUAGGCCACGACAAU__CUUGAGCAUUUGUUUGGCCACAAUAUGGCCAGGCCAUGGCACUUUUGACAACUUGACAGUCGUGCAGACAG____UC ...............((((.(((((.(((((...........(((((....))))).........))))))))))...(((....((((.........)))))))))))........ (-24.02 = -24.22 + 0.19)


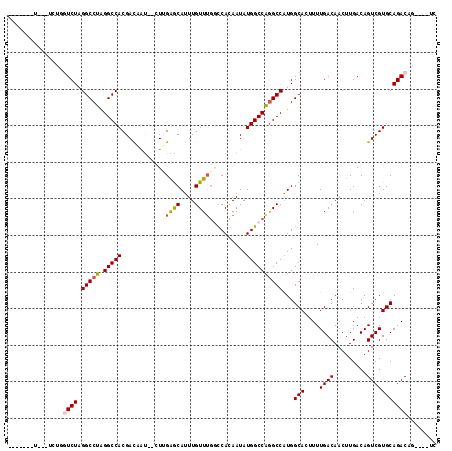
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:52 2006