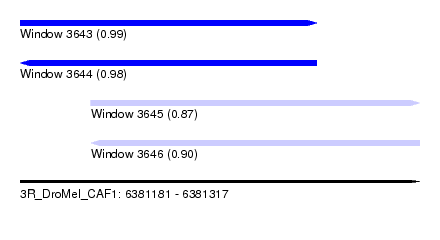
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,381,181 – 6,381,317 |
| Length | 136 |
| Max. P | 0.994191 |

| Location | 6,381,181 – 6,381,282 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 74.25 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -16.53 |
| Energy contribution | -16.53 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6381181 101 + 27905053 A-A--AGAAAUGAAAGGAACAAAAGUAUAAGUACGAAAUGGAACUUU-UGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUU-UUUCCAUUGAAAGCGUUAUAU .-.--...................((((((((....(((((((((((-(((.....((((((....))))))))))))).....-.)))))))....)).)))))) ( -24.70) >DroPse_CAF1 144034 87 + 1 -----GGCUGAGAAAC---CGAAA-U-------GGAAAUGGAACUUU-UGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUU-UUUCCAUUGAAAGCGUA-CGG -----.(((.......---...((-(-------(((((.....((((-(((.....((((((....))))))))))))).....-))))))))...)))...-... ( -26.22) >DroEre_CAF1 156398 101 + 1 A-U--AAGAAUGAAAGGAACGAGCGCAUAAGUGCGAAAUGGAACUUU-UGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUU-UUUCCAUUGAAAGCGUUAUAU .-.--............((((.((((....))))..(((((((((((-(((.....((((((....))))))))))))).....-.))))))).....)))).... ( -29.10) >DroWil_CAF1 241165 85 + 1 --------------AAAAAAAAA--AACCAA-ACAAAAUGGAACUUUUUCUGCCACGGCGCCAUUUGGCGCUACAA-AGAUAUUUUUUCCAUUGAAAGCAUA---A --------------.........--......-....(((((((.....(((.....((((((....))))))....-)))......))))))).........---. ( -19.50) >DroAna_CAF1 145482 100 + 1 UGUACGGACGCGAAAG---CGAAAACGCCAG-CCGAAAUGGAACUUU-UGCGCCAAGGCGCCAUUUGGCGCUACAAAAGAUAUU-UUUCCAUUGAAAGCAUUAUAU .....((.(((....)---))......)).(-(...(((((((((((-(((((((((......))))))))...))))).....-.)))))))....))....... ( -28.60) >DroPer_CAF1 151235 87 + 1 -----GGCUGAGAAAC---CGAAA-U-------GGAAAUGGAACUUU-UGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUU-UUUCCAUUGAAAGCGUA-CGG -----.(((.......---...((-(-------(((((.....((((-(((.....((((((....))))))))))))).....-))))))))...)))...-... ( -26.22) >consensus _____GGAAGAGAAAG___CGAAA_UA__AG__CGAAAUGGAACUUU_UGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUU_UUUCCAUUGAAAGCGUA_UAU ....................................(((((((.............((((((....))))))..............)))))))............. (-16.53 = -16.53 + -0.00)

| Location | 6,381,181 – 6,381,282 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 74.25 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -15.91 |
| Energy contribution | -15.91 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.978981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6381181 101 - 27905053 AUAUAACGCUUUCAAUGGAAA-AAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACA-AAAGUUCCAUUUCGUACUUAUACUUUUGUUCCUUUCAUUUCU--U-U .............(((((((.-.............(((((....)))))...((.(((-(((((................)))))))).)))))))))...--.-. ( -21.69) >DroPse_CAF1 144034 87 - 1 CCG-UACGCUUUCAAUGGAAA-AAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACA-AAAGUUCCAUUUCC-------A-UUUCG---GUUUCUCAGCC----- ...-.........((((((((-.....(((((((...(((....)))((....)))))-)))).....)))))-------)-))..(---((......)))----- ( -22.90) >DroEre_CAF1 156398 101 - 1 AUAUAACGCUUUCAAUGGAAA-AAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACA-AAAGUUCCAUUUCGCACUUAUGCGCUCGUUCCUUUCAUUCUU--A-U ....((((.....(((((((.-.....(((((((...(((....)))((....)))))-))))))))))).((((....))))..))))............--.-. ( -22.40) >DroWil_CAF1 241165 85 - 1 U---UAUGCUUUCAAUGGAAAAAAUAUCU-UUGUAGCGCCAAAUGGCGCCGUGGCAGAAAAAGUUCCAUUUUGU-UUGGUU--UUUUUUUUU-------------- .---...(((..((((((((......(((-(..(.(((((....))))).)..).))).....))))))..)).-..))).--.........-------------- ( -19.60) >DroAna_CAF1 145482 100 - 1 AUAUAAUGCUUUCAAUGGAAA-AAUAUCUUUUGUAGCGCCAAAUGGCGCCUUGGCGCA-AAAGUUCCAUUUCGG-CUGGCGUUUUCG---CUUUCGCGUCCGUACA .......(((...(((((((.-.....(((((((...(((....)))((....)))))-)))))))))))..))-)..(((....((---(....)))..)))... ( -26.50) >DroPer_CAF1 151235 87 - 1 CCG-UACGCUUUCAAUGGAAA-AAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACA-AAAGUUCCAUUUCC-------A-UUUCG---GUUUCUCAGCC----- ...-.........((((((((-.....(((((((...(((....)))((....)))))-)))).....)))))-------)-))..(---((......)))----- ( -22.90) >consensus AUA_AACGCUUUCAAUGGAAA_AAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACA_AAAGUUCCAUUUCG__CU__UA_UUUCG___CUUUCACAGCC_____ .............(((((((..........(((..(((((....)))))..))).........))))))).................................... (-15.91 = -15.91 + 0.00)


| Location | 6,381,205 – 6,381,317 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -14.77 |
| Energy contribution | -16.27 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
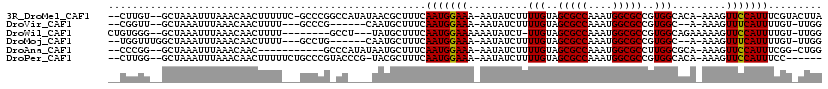
>3R_DroMel_CAF1 6381205 112 + 27905053 UAAGUACGAAAUGGAACUUU-UGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUU-UUUCCAUUGAAAGCGUUAUAUGGCCGGGC-GAAAAAGUUGUUUAAAUUUAGC--ACAAG-- (((((...((((..((((((-(.((((.((((..(((.(((((((.(((..((....-..))..)))..))))))).))))))))))-).)))))))))))..)))))..--.....-- ( -31.00) >DroVir_CAF1 155592 101 + 1 CCAA-ACAAAAUGAAACUUU-U--GCCACGGCGCCAUUUGGCGCUACAAAAGAUAUU-UUUCCAUUGAAAGCAUUG------CGGGC---AAAAGUUGUUUAAAUUUAGC--AACCG-- ....-.........((((((-(--(((.(((((((....)))))..(((..((....-..))..))).........------)))))---))))))).............--.....-- ( -26.70) >DroWil_CAF1 241176 104 + 1 CCAA-ACAAAAUGGAACUUUUUCUGCCACGGCGCCAUUUGGCGCUACAA-AGAUAUUUUUUCCAUUGAAAGCAUA---AGGC--------AAAAGUUGUUUAAAUUUAGC--CCCACAG ..((-(((((((((((.....(((.....((((((....))))))....-)))......)))))))....((...---..))--------.....)))))).........--....... ( -24.00) >DroMoj_CAF1 202325 103 + 1 CCAA-ACAAAAUGAAACUUU-U--GCCACGGCGCCAUUUGGCGCUACAAAAGAUAUU-UUUCCAUUGAAAGCAUUG------CAGGC---AAAAGUUGUUUAAAUUUAGCCAAACCA-- ....-.........((((((-(--(((..((((((....)))))).(((..((....-..))..))).........------..)))---)))))))....................-- ( -26.60) >DroAna_CAF1 145506 101 + 1 CCAG-CCGAAAUGGAACUUU-UGCGCCAAGGCGCCAUUUGGCGCUACAAAAGAUAUU-UUUCCAUUGAAAGCAUUAUAUGGGC-----------GUUGUUUAAAUUUAGC--CCGGG-- ((.(-(...(((((((((((-(((((((((......))))))))...))))).....-.)))))))....)).......((((-----------..............))--)))).-- ( -30.54) >DroPer_CAF1 151252 106 + 1 ------GGAAAUGGAACUUU-UGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUU-UUUCCAUUGAAAGCGUA-CGGGUACGGGCAGAAAAAGUUGUUUAAAUUUAGC--CCAAG-- ------(((((.....((((-(((.....((((((....))))))))))))).....-))))).........(((-....)))((((.....(((((.....))))).))--))...-- ( -29.90) >consensus CCAA_ACAAAAUGGAACUUU_UGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUU_UUUCCAUUGAAAGCAUU____GG_CGGGC___AAAAGUUGUUUAAAUUUAGC__ACCAG__ ..............((((((....(((..((((((....)))))).(((..((.......))..))).................)))....))))))...................... (-14.77 = -16.27 + 1.50)

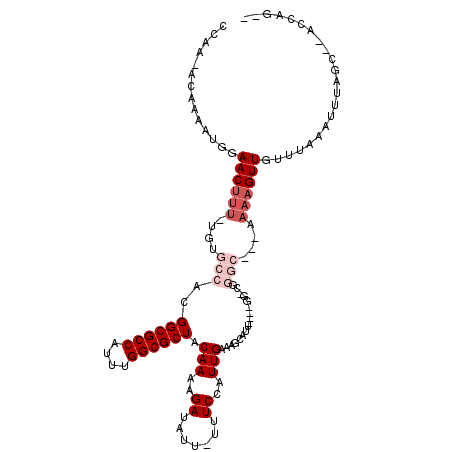
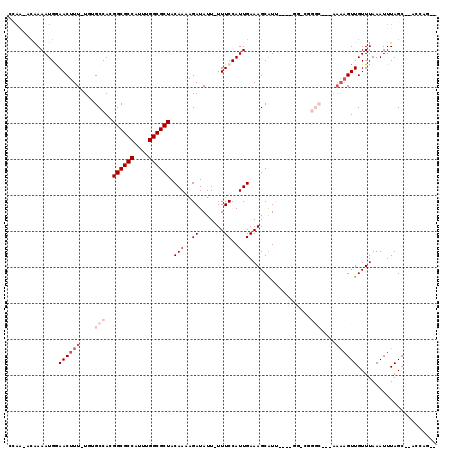
| Location | 6,381,205 – 6,381,317 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -15.16 |
| Energy contribution | -14.94 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6381205 112 - 27905053 --CUUGU--GCUAAAUUUAAACAACUUUUUC-GCCCGGCCAUAUAACGCUUUCAAUGGAAA-AAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACA-AAAGUUCCAUUUCGUACUUA --...((--((.((((......(((((((..-(((((((........(((..(((.(((..-....))).))).)))(((....))))))).)))..)-))))))..)))).))))... ( -31.40) >DroVir_CAF1 155592 101 - 1 --CGGUU--GCUAAAUUUAAACAACUUUU---GCCCG------CAAUGCUUUCAAUGGAAA-AAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGC--A-AAAGUUUCAUUUUGU-UUGG --.....--.((((((..((..(((((((---(((((------(...(((..(((.(((..-....))).))).)))(((....)))).)).)))--)-))))))...))..))-)))) ( -29.00) >DroWil_CAF1 241176 104 - 1 CUGUGGG--GCUAAAUUUAAACAACUUUU--------GCCU---UAUGCUUUCAAUGGAAAAAAUAUCU-UUGUAGCGCCAAAUGGCGCCGUGGCAGAAAAAGUUCCAUUUUGU-UUGG ..(((((--((..................--------))))---)))......(((((((......(((-(..(.(((((....))))).)..).))).....)))))))....-.... ( -26.27) >DroMoj_CAF1 202325 103 - 1 --UGGUUUGGCUAAAUUUAAACAACUUUU---GCCUG------CAAUGCUUUCAAUGGAAA-AAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGC--A-AAAGUUUCAUUUUGU-UUGG --........((((((..((..(((((((---(((.(------(...(((..(((.(((..-....))).))).)))(((....)))))...)))--)-))))))...))..))-)))) ( -28.90) >DroAna_CAF1 145506 101 - 1 --CCCGG--GCUAAAUUUAAACAAC-----------GCCCAUAUAAUGCUUUCAAUGGAAA-AAUAUCUUUUGUAGCGCCAAAUGGCGCCUUGGCGCA-AAAGUUCCAUUUCGG-CUGG --.((((--((..............-----------)))).......(((...(((((((.-.....(((((((...(((....)))((....)))))-)))))))))))..))-).)) ( -28.94) >DroPer_CAF1 151252 106 - 1 --CUUGG--GCUAAAUUUAAACAACUUUUUCUGCCCGUACCCG-UACGCUUUCAAUGGAAA-AAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACA-AAAGUUCCAUUUCC------ --...((--((.....................))))(((....-)))......(((((((.-.....(((((((...(((....)))((....)))))-)))))))))))...------ ( -26.10) >consensus __CUGGG__GCUAAAUUUAAACAACUUUU___GCCCG_CC____AAUGCUUUCAAUGGAAA_AAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACA_AAAGUUCCAUUUCGU_UUGG .....................................................(((((((..........(((..(((((....)))))..))).........)))))))......... (-15.16 = -14.94 + -0.22)
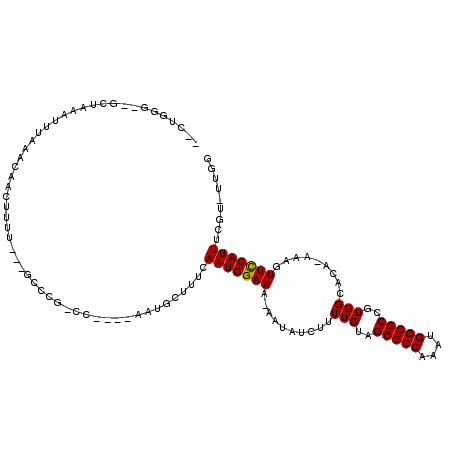
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:49 2006