
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 888,820 – 888,956 |
| Length | 136 |
| Max. P | 0.999919 |

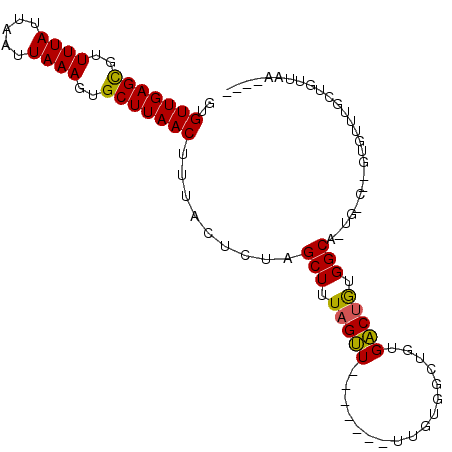
| Location | 888,820 – 888,922 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 74.07 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -16.21 |
| Energy contribution | -15.66 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.61 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 888820 102 - 27905053 GUGUUGAGUGUUUUAUUAAUUAAAGUGCUUAACU-CACUAUGGCUAUUGCUAUUGCUAUGAUGGCUAUGGCUAUGGCUUUGGCUCUUGGUUGCAGUUGUUGUU (.((((((..(((((.....)))))..)))))).-)((((.(((((..(((((.((((((.....)))))).)))))..)))))..))))............. ( -33.10) >DroPse_CAF1 64541 91 - 1 GUGUUGAGCGUUUUAUUAAUUAAAGUGCUUAACUUUACUCUAGCUUUAGUU-------UUGUGGCUGUGACUGUGGCAGUGACA-GUGUUUGCUGUUAA---- ..(((((((((((((.....))))))))))))).........(((.(((((-------..........))))).)))..(((((-((....))))))).---- ( -24.80) >DroPer_CAF1 64420 85 - 1 GUGUUGAGCGUUUUAUUAAUUAAAGUGCUUAACUUUACUCUAGCUUUAGUU-------UUGUGGCUGUGACUGUGGCA-------GUGUUUGCUGUUAA---- ..(((((((((((((.....))))))))))))).....(((((((......-------....))))).))...(((((-------((....))))))).---- ( -21.40) >consensus GUGUUGAGCGUUUUAUUAAUUAAAGUGCUUAACUUUACUCUAGCUUUAGUU_______UUGUGGCUGUGACUGUGGCA_UG_C__GUGUUUGCUGUUAA____ ..(((((((..((((.....))))..))))))).........(((.(((((.................))))).))).......................... (-16.21 = -15.66 + -0.55)


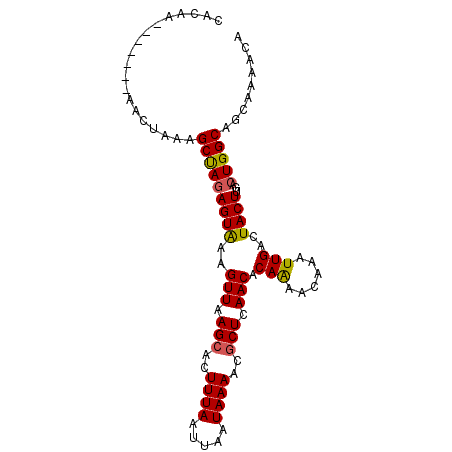
| Location | 888,860 – 888,956 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 86.62 |
| Mean single sequence MFE | -12.13 |
| Consensus MFE | -9.83 |
| Energy contribution | -9.83 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 888860 96 + 27905053 CAUCAUAGCAAUAGCAAUAGCCAUAGUG-AGUUAAGCACUUUAAUUAAUAAAACACUCAACACAGAACAAAUUGACUACUUGACUGGCAGCAACACA .......((....((....))....((.-(((((((....((((((.......................))))))...))))))).)).))...... ( -12.20) >DroPse_CAF1 64576 90 + 1 CACAA-------AACUAAAGCUAGAGUAAAGUUAAGCACUUUAAUUAAUAAAACGCUCAACACAAAACAAAUUGACUACUUGACUGGCAGCAAAACA .....-------.......(((((((((..(((.(((..((((.....))))..))).))).(((......)))..))))...)))))......... ( -12.10) >DroPer_CAF1 64449 90 + 1 CACAA-------AACUAAAGCUAGAGUAAAGUUAAGCACUUUAAUUAAUAAAACGCUCAACACAAAACAAAUUGACUACUUGACUGGCAGCAAAACA .....-------.......(((((((((..(((.(((..((((.....))))..))).))).(((......)))..))))...)))))......... ( -12.10) >consensus CACAA_______AACUAAAGCUAGAGUAAAGUUAAGCACUUUAAUUAAUAAAACGCUCAACACAAAACAAAUUGACUACUUGACUGGCAGCAAAACA ...................(((((((((..(((.(((..((((.....))))..))).))).(((......)))..))))...)))))......... ( -9.83 = -9.83 + 0.01)

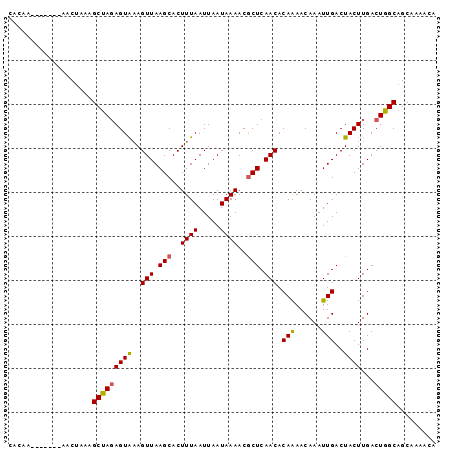
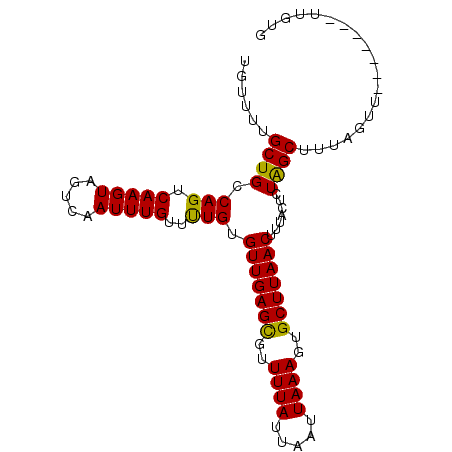
| Location | 888,860 – 888,956 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.62 |
| Mean single sequence MFE | -17.97 |
| Consensus MFE | -16.63 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 888860 96 - 27905053 UGUGUUGCUGCCAGUCAAGUAGUCAAUUUGUUCUGUGUUGAGUGUUUUAUUAAUUAAAGUGCUUAACU-CACUAUGGCUAUUGCUAUUGCUAUGAUG ...((((((((.......)))).))))......((.((((((..(((((.....)))))..)))))).-)).((((((..........))))))... ( -21.70) >DroPse_CAF1 64576 90 - 1 UGUUUUGCUGCCAGUCAAGUAGUCAAUUUGUUUUGUGUUGAGCGUUUUAUUAAUUAAAGUGCUUAACUUUACUCUAGCUUUAGUU-------UUGUG ......((((.(((.(((((.....)))))..))).(((((((((((((.....))))))))))))).......)))).......-------..... ( -16.10) >DroPer_CAF1 64449 90 - 1 UGUUUUGCUGCCAGUCAAGUAGUCAAUUUGUUUUGUGUUGAGCGUUUUAUUAAUUAAAGUGCUUAACUUUACUCUAGCUUUAGUU-------UUGUG ......((((.(((.(((((.....)))))..))).(((((((((((((.....))))))))))))).......)))).......-------..... ( -16.10) >consensus UGUUUUGCUGCCAGUCAAGUAGUCAAUUUGUUUUGUGUUGAGCGUUUUAUUAAUUAAAGUGCUUAACUUUACUCUAGCUUUAGUU_______UUGUG ......((((.(((.(((((.....)))))..))).(((((((..((((.....))))..))))))).......))))................... (-16.63 = -15.97 + -0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:05 2006