
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,344,346 – 6,344,439 |
| Length | 93 |
| Max. P | 0.961636 |

| Location | 6,344,346 – 6,344,439 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.69 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
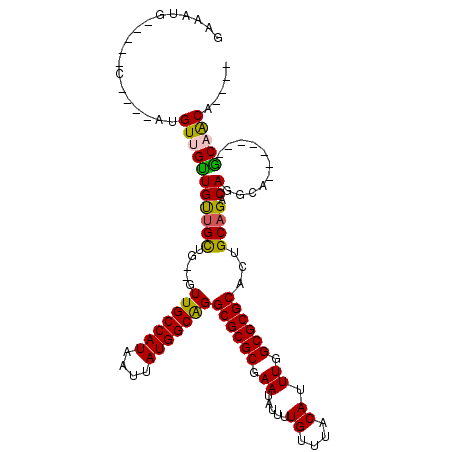
>3R_DroMel_CAF1 6344346 93 + 27905053 ----UGUUGC-------UGCCUGUCUGCAGUGCGCGCCAAAUGUAAACAAAAUAUUCGCGCGCCCGCCAUAAUUAUGGCAAC--CAGCAACAACAACAU----G-----CAUUUC ----((((((-------((.(((....))).((((((..(((((.......))))).))))))..(((((....)))))...--)))))))).......----.-----...... ( -31.20) >DroSim_CAF1 122687 93 + 1 ----UGUAGC-------UGCCUGUCUGCAGUGCGCGCCAAAUGUAAACAAAAUAUUCGCGCGCCUGCCAUAAUUAUGGCAAC--CAGCAACAACAACAU----G-----CAUUUC ----(((.((-------((..((((.((((.((((((..(((((.......))))).)))))))))).........))))..--)))).))).......----.-----...... ( -28.80) >DroEre_CAF1 120283 98 + 1 ----UGUUGC-------UGCCUGUCUGCAGUGCGCGCCAAAUGUAAACAAAAUAUUCGCGCGCCUGCCAUAAUUAUGGCAAC--CAGCUACAACAACAU----GCAAUUCUGUUC ----(((.((-------((..((((.((((.((((((..(((((.......))))).)))))))))).........))))..--)))).))).......----............ ( -28.80) >DroWil_CAF1 198709 87 + 1 ----UGCUGU-------UGGCUGUCUGCAAUGCGCGCCAAAUGUAAACAAAAUAUUUGCGCGCCUGCCAUAAUUAUGGCAACUCCAACAACAACAACA----------------- ----((.(((-------(((..((.......((((((.((((((.......)))))))))))).((((((....)))))))).)))))).))......----------------- ( -29.20) >DroYak_CAF1 109858 106 + 1 CUGUUGUUGC-------UGCCUGUCUGCAGUGCGCGCCAAAUGUAAACAAAAUAUUCGCGCGCCUGCCAUAAUUAUGGCAAC--CAGCAACAGCAACAACAAGGACAUUCAUUUC .(((((((((-------((..((.(((.((.((((((..(((((.......))))).))))))))(((((....)))))...--))))).))))))))))).............. ( -39.60) >DroAna_CAF1 111171 91 + 1 ----UGCAGUUCUGGCGUGGCUGUCUGCAGUGCGCGCCAAAUGUAAACAAAAUAUUCGCGCGCCUGCCAUAAUUAUGGCAAC--CGGCAGCAACAAC------------------ ----((((....(((((((((((....)))).)))))))..))))............((..(((((((((....))))))..--.))).))......------------------ ( -33.40) >consensus ____UGUUGC_______UGCCUGUCUGCAGUGCGCGCCAAAUGUAAACAAAAUAUUCGCGCGCCUGCCAUAAUUAUGGCAAC__CAGCAACAACAACAU____G_____CAUUUC ....(((.((.......((((.....((((.((((((..(((((.......))))).)))))))))).........))))......)).)))....................... (-21.44 = -21.69 + 0.25)



| Location | 6,344,346 – 6,344,439 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.10 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6344346 93 - 27905053 GAAAUG-----C----AUGUUGUUGUUGCUG--GUUGCCAUAAUUAUGGCGGGCGCGCGAAUAUUUUGUUUACAUUUGGCGCGCACUGCAGACAGGCA-------GCAACA---- ......-----.----....((((((((((.--((((((((....)))))((((((((.((.....((....)).)).)))))).))...))).))))-------))))))---- ( -35.90) >DroSim_CAF1 122687 93 - 1 GAAAUG-----C----AUGUUGUUGUUGCUG--GUUGCCAUAAUUAUGGCAGGCGCGCGAAUAUUUUGUUUACAUUUGGCGCGCACUGCAGACAGGCA-------GCUACA---- ......-----.----.(((.((((((.(((--.(((((((....)))))))((((((.((.....((....)).)).))))))....)))...))))-------)).)))---- ( -32.10) >DroEre_CAF1 120283 98 - 1 GAACAGAAUUGC----AUGUUGUUGUAGCUG--GUUGCCAUAAUUAUGGCAGGCGCGCGAAUAUUUUGUUUACAUUUGGCGCGCACUGCAGACAGGCA-------GCAACA---- ............----.(((((((((..(((--.(((((((....)))))))((((((.((.....((....)).)).))))))....)))....)))-------))))))---- ( -35.40) >DroWil_CAF1 198709 87 - 1 -----------------UGUUGUUGUUGUUGGAGUUGCCAUAAUUAUGGCAGGCGCGCAAAUAUUUUGUUUACAUUUGGCGCGCAUUGCAGACAGCCA-------ACAGCA---- -----------------.((((((((((((....(((((((....)))))))((((((((((...........)))).))))))......))))).))-------))))).---- ( -32.50) >DroYak_CAF1 109858 106 - 1 GAAAUGAAUGUCCUUGUUGUUGCUGUUGCUG--GUUGCCAUAAUUAUGGCAGGCGCGCGAAUAUUUUGUUUACAUUUGGCGCGCACUGCAGACAGGCA-------GCAACAACAG ..............(((((((((((((.(((--.(((((((....)))))))((((((.((.....((....)).)).))))))....)))...))))-------))))))))). ( -44.50) >DroAna_CAF1 111171 91 - 1 ------------------GUUGUUGCUGCCG--GUUGCCAUAAUUAUGGCAGGCGCGCGAAUAUUUUGUUUACAUUUGGCGCGCACUGCAGACAGCCACGCCAGAACUGCA---- ------------------((.((((((((.(--.(((((((....)))))))((((((.((.....((....)).)).)))))).).)))).)))).))((.......)).---- ( -33.30) >consensus GAAAUG_____C____AUGUUGUUGUUGCUG__GUUGCCAUAAUUAUGGCAGGCGCGCGAAUAUUUUGUUUACAUUUGGCGCGCACUGCAGACAGGCA_______GCAACA____ ..................(((((((((((.....(((((((....)))))))((((((.((.....((....)).)).))))))...)))).))...........)))))..... (-26.60 = -26.10 + -0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:24 2006