
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,338,384 – 6,338,521 |
| Length | 137 |
| Max. P | 0.747398 |

| Location | 6,338,384 – 6,338,484 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.60 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6338384 100 + 27905053 --------UUUGACUUUUCAAUUUGCAGUUCAGUUCGUUUGCGAUCUGCCCCUGAUUUUUG-CUCAUUUUGGGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCA --------.((((....)))).......(((((..((((.((((((((((((.........-........)))))))).....))))))))..)))))........... ( -27.33) >DroVir_CAF1 109191 92 + 1 UA-----UUUUUUUUUUUCAAUUUGCAGUUCAUUUCUGCUGC----------UGG-CUUUG-CUCAUUUUGAGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCA ..-----........(((((....(((((........)))))----------.((-(.(((-(.((.(((.......))).))..)))).))).))))).......... ( -18.00) >DroPse_CAF1 100247 99 + 1 --UUGGACUUGGACUUUUCAAUUUGCUGUUGAUUUCUGCUGUGUUC-------GAGCUCUG-CUCAUUUUAGGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCA --..((((.(((.....(((((.....)))))......))).))))-------((((...)-)))(((((((((((.............)))))))))))......... ( -26.82) >DroGri_CAF1 130418 94 + 1 CUU---GAUUUUUUUUUUCAAUUUGCAGUUCAUUUCUGUUGC----------UGG-CUGUG-CUCAUUUUGAGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCA ..(---(((......(((((..((((((...((((((((.((----------...-..)).-(((.....)))))))))))..)))))).....)))))......)))) ( -17.40) >DroAna_CAF1 105388 105 + 1 -CU---UUUUUGACUUUUCAAUUUGCAGUUCGUUUGGACUGUCCGGUGGCCCCAAUUGCUGCCUCAUUUUGGGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCA -..---.....(((.(((((....(((((((....)))))))..((((((..(((((.(((((((.....))))))).)))))..))..)))).)))))..)))..... ( -31.00) >DroPer_CAF1 106581 93 + 1 --------UUGGACUUUUCAAUUUGCUGUUGAUUUCUGCUGUGUUC-------GAGCUCUG-CUCAUUUUAGGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCA --------.(((((.(((((..((((...(((((((((((.((...-------((((...)-)))....)).)))))))))))..)))).....)))))..)))))... ( -25.00) >consensus ________UUUGACUUUUCAAUUUGCAGUUCAUUUCUGCUGCG_UC______UGA_CUCUG_CUCAUUUUGGGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCA ........................(((((........))))).......................(((((((((((.............)))))))))))......... (-14.57 = -14.60 + 0.03)

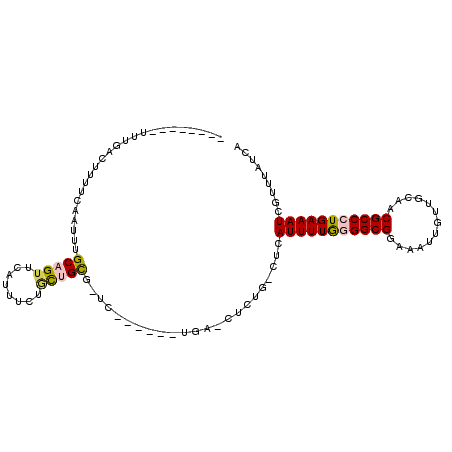

| Location | 6,338,415 – 6,338,521 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -21.41 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6338415 106 + 27905053 UGCGAUCUGCCCCUGAUUUUUG-CUCAUUUUGGGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCAAAACGACAAAAUGGAAUUGAGUUCCGUUCUGCCAUA--U (((((((((((((.........-........)))))))).....)))))...........((((((....))))))((.((((((((...)))))))).)).....--. ( -28.43) >DroVir_CAF1 109225 88 + 1 UGC----------UGG-CUUUG-CUCAUUUUGAGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCAAAACGACAAAAUGGAAUUGAGUUCCGUUGC--------- ...----------.((-(.(((-(.((.(((.......))).))..)))).)))......((((((....))))))...((((((((...))))))))..--------- ( -19.10) >DroPse_CAF1 100284 93 + 1 UGUGUUC-------GAGCUCUG-CUCAUUUUAGGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCAAAACGACAAAAUGGAAUUGAGUUCCGUU--GCC------ .......-------((((...)-))).((((((((((.............))))))))))((((((....))))))...((((((((...))))))))--...------ ( -28.82) >DroGri_CAF1 130454 88 + 1 UGC----------UGG-CUGUG-CUCAUUUUGAGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCAAAACGACAAAAUGGAAUUGAGUUCCGUUGC--------- ...----------...-((((.-(((.....)))))))........(((((.........((((((....))))))......(((((...))))))))))--------- ( -19.20) >DroAna_CAF1 105423 109 + 1 UGUCCGGUGGCCCCAAUUGCUGCCUCAUUUUGGGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCAAAACGACAAAAUGGAAUCGAGUUCCGUUCCACCAUGCCC .....((((((..(((((.(((((((.....))))))).)))))..))............((((((....))))))...((((((((...)))))))).))))...... ( -32.90) >DroPer_CAF1 106612 93 + 1 UGUGUUC-------GAGCUCUG-CUCAUUUUAGGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCAAAACGACAAAAUGGAAUUGAGUUCCGUU--GCC------ .......-------((((...)-))).((((((((((.............))))))))))((((((....))))))...((((((((...))))))))--...------ ( -28.82) >consensus UGCG_UC______UGA_CUCUG_CUCAUUUUGGGGCGGAAAUUGUUGCAACGCCCUGAAAUCGUUUAUCAAAACGACAAAAUGGAAUUGAGUUCCGUU_CGCC______ ...........................((((((((((.............))))))))))((((((....))))))...((((((((...))))))))........... (-21.41 = -21.52 + 0.11)



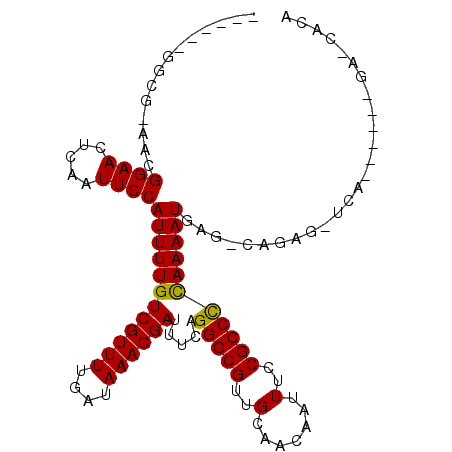
| Location | 6,338,415 – 6,338,521 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -16.94 |
| Energy contribution | -16.50 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6338415 106 - 27905053 A--UAUGGCAGAACGGAACUCAAUUCCAUUUUGUCGUUUUGAUAAACGAUUUCAGGGCGUUGCAACAAUUUCCGCCCCAAAAUGAG-CAAAAAUCAGGGGCAGAUCGCA .--.(((((((((.((((.....)))).))))))))).........((((((..(((.((((...)))).)))(((((....(((.-......)))))))))))))).. ( -30.10) >DroVir_CAF1 109225 88 - 1 ---------GCAACGGAACUCAAUUCCAUUUUGUCGUUUUGAUAAACGAUUUCAGGGCGUUGCAACAAUUUCCGCCUCAAAAUGAG-CAAAG-CCA----------GCA ---------((...((((.....)))).((((((((((((((.....(....)..((((..(.......)..))))))))))))).-)))))-...----------)). ( -19.80) >DroPse_CAF1 100284 93 - 1 ------GGC--AACGGAACUCAAUUCCAUUUUGUCGUUUUGAUAAACGAUUUCAGGGCGUUGCAACAAUUUCCGCCCUAAAAUGAG-CAGAGCUC-------GAACACA ------(..--..)((((.....)))).....(((((((....)))))))(((((((((..(.......)..)))))).....(((-(...))))-------))).... ( -23.40) >DroGri_CAF1 130454 88 - 1 ---------GCAACGGAACUCAAUUCCAUUUUGUCGUUUUGAUAAACGAUUUCAGGGCGUUGCAACAAUUUCCGCCUCAAAAUGAG-CACAG-CCA----------GCA ---------((..(((((..((((.((.....(((((((....))))))).....)).)))).......))))).(((.....)))-.....-...----------)). ( -18.70) >DroAna_CAF1 105423 109 - 1 GGGCAUGGUGGAACGGAACUCGAUUCCAUUUUGUCGUUUUGAUAAACGAUUUCAGGGCGUUGCAACAAUUUCCGCCCCAAAAUGAGGCAGCAAUUGGGGCCACCGGACA ..(..((((((..(((((..((((.((.....(((((((....))))))).....)).)))).......)))))((((((..((......)).))))))))))))..). ( -32.30) >DroPer_CAF1 106612 93 - 1 ------GGC--AACGGAACUCAAUUCCAUUUUGUCGUUUUGAUAAACGAUUUCAGGGCGUUGCAACAAUUUCCGCCCUAAAAUGAG-CAGAGCUC-------GAACACA ------(..--..)((((.....)))).....(((((((....)))))))(((((((((..(.......)..)))))).....(((-(...))))-------))).... ( -23.40) >consensus ______GGCG_AACGGAACUCAAUUCCAUUUUGUCGUUUUGAUAAACGAUUUCAGGGCGUUGCAACAAUUUCCGCCCCAAAAUGAG_CAGAG_UCA______GA_CACA ..............((((.....))))((((((((((((....)))))).....(((((..(.......)..))))))))))).......................... (-16.94 = -16.50 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:18 2006