
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,333,834 – 6,333,956 |
| Length | 122 |
| Max. P | 0.996849 |

| Location | 6,333,834 – 6,333,938 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -19.48 |
| Consensus MFE | -17.81 |
| Energy contribution | -18.38 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
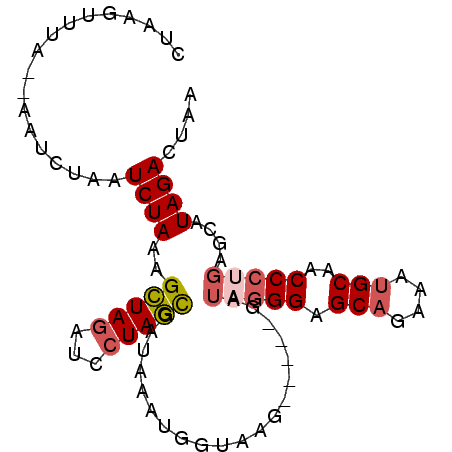
>3R_DroMel_CAF1 6333834 104 + 27905053 CAAGUCCUUUGUUAAUUU----UUUUCUGGUGCAUUUUAGUCUAUGCUCAGGGUUGCAUUUCUGCUCCCUAUCCGUCUCUUACCAUUUAUCGCUAGGAUCUAGCUUUA ..................----.....((((((((........))))..((((..(((....))).))))...........))))......(((((...))))).... ( -19.50) >DroSim_CAF1 112184 104 + 1 CAUUUCCAUUGUUAAUUUUCUUUUUUCUGGUGCAUUUUAGUCUAUGCUCGGGGUUGCAUUUCUGCUCCCUAUCC----CUUACCAUUUAUCGCUAGGAUCUAGCUUUA ...........................((((((((........))))..((((..(((....))).))))....----...))))......(((((...))))).... ( -19.50) >DroEre_CAF1 109773 102 + 1 CAUGUCCUUUGUUAAUUU-----UUUCUGGUGCACUUUAGUCUAUGCUCAGGGUUGCAUUUCUGCGCCCUAUC-GUCCCUUACCAUUUAUCGCUAGGAUCUAGCUUUA ..................-----....(((((((..........)))..(((((.(((....))))))))...-.......))))......(((((...))))).... ( -18.10) >DroYak_CAF1 99068 103 + 1 CAUGUCCUUUGUUAAUUUA----CUUCUGGUGCAUUUUAGUCUAUGCUCAGGGUUGCAUUUCUGCGCCCUAUC-GUCCCUUACCAUUUAUCGCUAGGAUCUAGCUUUA ...................----....((((((((........))))..(((((.(((....))))))))...-.......))))......(((((...))))).... ( -20.80) >consensus CAUGUCCUUUGUUAAUUU_____UUUCUGGUGCAUUUUAGUCUAUGCUCAGGGUUGCAUUUCUGCGCCCUAUC_GUCCCUUACCAUUUAUCGCUAGGAUCUAGCUUUA ...........................((((((((........))))..(((((.(((....))))))))...........))))......(((((...))))).... (-17.81 = -18.38 + 0.56)



| Location | 6,333,834 – 6,333,938 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6333834 104 - 27905053 UAAAGCUAGAUCCUAGCGAUAAAUGGUAAGAGACGGAUAGGGAGCAGAAAUGCAACCCUGAGCAUAGACUAAAAUGCACCAGAAAA----AAAUUAACAAAGGACUUG ....(((((...)))))......((((..........(((((.(((....)))..))))).((((........)))))))).....----.................. ( -23.40) >DroSim_CAF1 112184 104 - 1 UAAAGCUAGAUCCUAGCGAUAAAUGGUAAG----GGAUAGGGAGCAGAAAUGCAACCCCGAGCAUAGACUAAAAUGCACCAGAAAAAAGAAAAUUAACAAUGGAAAUG ....(((((...)))))......((((...----.....(((.(((....)))...)))..((((........))))))))........................... ( -21.40) >DroEre_CAF1 109773 102 - 1 UAAAGCUAGAUCCUAGCGAUAAAUGGUAAGGGAC-GAUAGGGCGCAGAAAUGCAACCCUGAGCAUAGACUAAAGUGCACCAGAAA-----AAAUUAACAAAGGACAUG ....(((((...)))))......((((.......-..(((((.(((....)))..))))).((((........))))))))....-----.................. ( -23.90) >DroYak_CAF1 99068 103 - 1 UAAAGCUAGAUCCUAGCGAUAAAUGGUAAGGGAC-GAUAGGGCGCAGAAAUGCAACCCUGAGCAUAGACUAAAAUGCACCAGAAG----UAAAUUAACAAAGGACAUG ....(((((...)))))......((((.......-..(((((.(((....)))..))))).((((........))))))))....----................... ( -23.60) >consensus UAAAGCUAGAUCCUAGCGAUAAAUGGUAAGGGAC_GAUAGGGAGCAGAAAUGCAACCCUGAGCAUAGACUAAAAUGCACCAGAAA_____AAAUUAACAAAGGACAUG ....(((((...)))))......((((..........(((((.(((....)))..))))).((((........))))))))........................... (-22.74 = -22.80 + 0.06)



| Location | 6,333,866 – 6,333,956 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -17.33 |
| Consensus MFE | -14.61 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6333866 90 + 27905053 UUAGUCUAUGCUCAGGGUUGCAUUUCUGCUCCCUAUCCGUCUCUUACCAUUUAUCGCUAGGAUCUAGCUUUAGAUUAGAUU--UAAACUUAA ((((((((.....((((..(((....))).)))).....................(((((...)))))..))))))))...--......... ( -18.60) >DroSec_CAF1 98661 86 + 1 UUAGUCUAUGCUCAGGGUUGCAUUUCUGCUCCCUAUCC----CUUACCAUUUAUCGCUAGGAUCUAGCUUUAGAUUAGAUU--UUAACUUAG ((((((((.....((((..(((....))).))))....----.............(((((...)))))..))))))))...--......... ( -18.60) >DroSim_CAF1 112220 86 + 1 UUAGUCUAUGCUCGGGGUUGCAUUUCUGCUCCCUAUCC----CUUACCAUUUAUCGCUAGGAUCUAGCUUUAGAUUAGAUU--UAAACUUAG ((((((((.....((((..(((....))).))))....----.............(((((...)))))..))))))))...--......... ( -18.60) >DroEre_CAF1 109804 89 + 1 UUAGUCUAUGCUCAGGGUUGCAUUUCUGCGCCCUAUC-GUCCCUUACCAUUUAUCGCUAGGAUCUAGCUUUAGAUUAGGUU--UAAACUUAG ((((((((.....(((((.(((....))))))))...-.................(((((...)))))..))))))))...--......... ( -19.90) >DroWil_CAF1 187956 74 + 1 UUAGUCUAUGCUCAGGGUUGCU--UUUGCUCC---------------GCUUUUUCAUUAUCAACUAACAUUAGAUUAAAUUAUUAUAUUUC- ((((((((.......(((((.(--..((....---------------.......))..).))))).....)))))))).............- ( -8.40) >DroYak_CAF1 99100 89 + 1 UUAGUCUAUGCUCAGGGUUGCAUUUCUGCGCCCUAUC-GUCCCUUACCAUUUAUCGCUAGGAUCUAGCUUUAGAUUAGGUU--UAAACUUAG ((((((((.....(((((.(((....))))))))...-.................(((((...)))))..))))))))...--......... ( -19.90) >consensus UUAGUCUAUGCUCAGGGUUGCAUUUCUGCUCCCUAUC_____CUUACCAUUUAUCGCUAGGAUCUAGCUUUAGAUUAGAUU__UAAACUUAG ((((((((.....((((..((......)).)))).....................(((((...)))))..)))))))).............. (-14.61 = -14.75 + 0.14)



| Location | 6,333,866 – 6,333,956 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -13.63 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6333866 90 - 27905053 UUAAGUUUA--AAUCUAAUCUAAAGCUAGAUCCUAGCGAUAAAUGGUAAGAGACGGAUAGGGAGCAGAAAUGCAACCCUGAGCAUAGACUAA ...((((((--..(((.(((....(((((...))))).......))).)))......(((((.(((....)))..)))))....)))))).. ( -22.00) >DroSec_CAF1 98661 86 - 1 CUAAGUUAA--AAUCUAAUCUAAAGCUAGAUCCUAGCGAUAAAUGGUAAG----GGAUAGGGAGCAGAAAUGCAACCCUGAGCAUAGACUAA (((.(((..--..(((.(((....(((((...))))).......))).))----)..(((((.(((....)))..)))))))).)))..... ( -19.60) >DroSim_CAF1 112220 86 - 1 CUAAGUUUA--AAUCUAAUCUAAAGCUAGAUCCUAGCGAUAAAUGGUAAG----GGAUAGGGAGCAGAAAUGCAACCCCGAGCAUAGACUAA ...((((((--..((..((((...((((.(((.....)))...))))...----)))).(((.(((....)))...)))))...)))))).. ( -19.80) >DroEre_CAF1 109804 89 - 1 CUAAGUUUA--AACCUAAUCUAAAGCUAGAUCCUAGCGAUAAAUGGUAAGGGAC-GAUAGGGCGCAGAAAUGCAACCCUGAGCAUAGACUAA ...((((((--..(((.(((....(((((...))))).......))).)))...-..(((((.(((....)))..)))))....)))))).. ( -24.30) >DroWil_CAF1 187956 74 - 1 -GAAAUAUAAUAAUUUAAUCUAAUGUUAGUUGAUAAUGAAAAAGC---------------GGAGCAAA--AGCAACCCUGAGCAUAGACUAA -........................((((((....(((....((.---------------((.((...--.))..))))...))).)))))) ( -7.60) >DroYak_CAF1 99100 89 - 1 CUAAGUUUA--AACCUAAUCUAAAGCUAGAUCCUAGCGAUAAAUGGUAAGGGAC-GAUAGGGCGCAGAAAUGCAACCCUGAGCAUAGACUAA ...((((((--..(((.(((....(((((...))))).......))).)))...-..(((((.(((....)))..)))))....)))))).. ( -24.30) >consensus CUAAGUUUA__AAUCUAAUCUAAAGCUAGAUCCUAGCGAUAAAUGGUAAG_____GAUAGGGAGCAGAAAUGCAACCCUGAGCAUAGACUAA ..................((((..(((((...)))))....................(((((.(((....)))..)))))....)))).... (-13.63 = -14.22 + 0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:13 2006