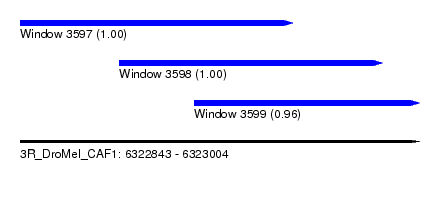
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,322,843 – 6,323,004 |
| Length | 161 |
| Max. P | 0.999970 |

| Location | 6,322,843 – 6,322,953 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 98.55 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -27.22 |
| Energy contribution | -27.22 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6322843 110 + 27905053 GUCGUCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCU (((((....))).)).........((((((..(((((........)))))...).)))))..(((((.(((.....(((((....)))))))).)))))........... ( -25.60) >DroPse_CAF1 83824 110 + 1 AGCGUCCUUGCGUACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCU .((((....))))...........((((((..(((((........)))))...).)))))..(((((.(((.....(((((....)))))))).)))))........... ( -26.60) >DroGri_CAF1 112648 110 + 1 CGCGUCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCU .((((....))))...........((((((..(((((........)))))...).)))))..(((((.(((.....(((((....)))))))).)))))........... ( -29.00) >DroWil_CAF1 172646 110 + 1 CGCGUCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCU .((((....))))...........((((((..(((((........)))))...).)))))..(((((.(((.....(((((....)))))))).)))))........... ( -29.00) >DroMoj_CAF1 135788 110 + 1 CGCGUCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCU .((((....))))...........((((((..(((((........)))))...).)))))..(((((.(((.....(((((....)))))))).)))))........... ( -29.00) >DroPer_CAF1 90106 110 + 1 AGCGUCCUUGCGUACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCU .((((....))))...........((((((..(((((........)))))...).)))))..(((((.(((.....(((((....)))))))).)))))........... ( -26.60) >consensus CGCGUCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCU .((((....))))...........((((((..(((((........)))))...).)))))..(((((.(((.....(((((....)))))))).)))))........... (-27.22 = -27.22 + -0.00)

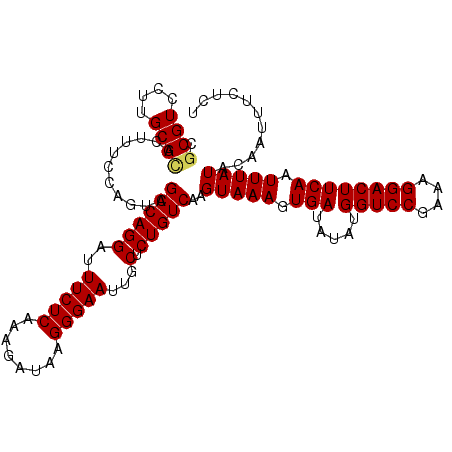
| Location | 6,322,883 – 6,322,989 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 95.13 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -25.05 |
| Energy contribution | -25.05 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.04 |
| SVM RNA-class probability | 0.999970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6322883 106 + 27905053 GAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU--UUU-CGCUACUUUUUGCGUUUGCUUUUUUUCC (((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))).--...-(((........))).............. ( -25.00) >DroVir_CAF1 93510 106 + 1 GAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU--UUUCCGCUACUUUUUGCGUUUUUCUG-UAUGC (((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))).--....(((........)))........-..... ( -25.20) >DroGri_CAF1 112688 106 + 1 GAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU--UUUCCGCUACUUUUUGCGUUUUUCUG-UAUGC (((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))).--....(((........)))........-..... ( -25.20) >DroEre_CAF1 94884 104 + 1 GAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU--UUU-CGCUACUUUUUGCGUUUGCUUU--UUCC (((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))).--...-(((........)))........--.... ( -25.00) >DroWil_CAF1 172686 107 + 1 GAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUUU-CGCUACUUUUUGCGUUUUUCUG-UUUGC (((((((((((((((......))))).((((.....(((((....))))).....)))).....))))))))))......-(((........)))........-..... ( -25.00) >DroMoj_CAF1 135828 106 + 1 GAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU--UUUCCGCUACUUUUUGCGUUUUUCUG-UAUGC (((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))).--....(((........)))........-..... ( -25.20) >consensus GAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU__UUU_CGCUACUUUUUGCGUUUUUCUG_UAUGC (((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))).......(((........))).............. (-25.05 = -25.05 + 0.00)

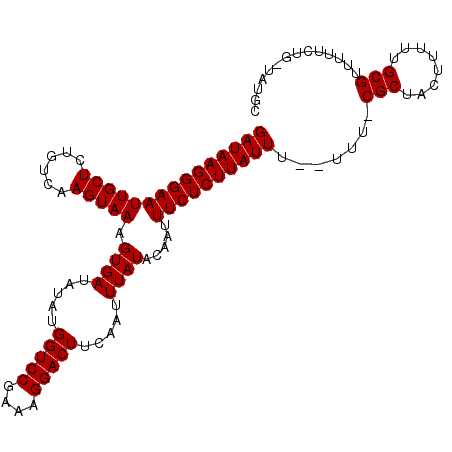
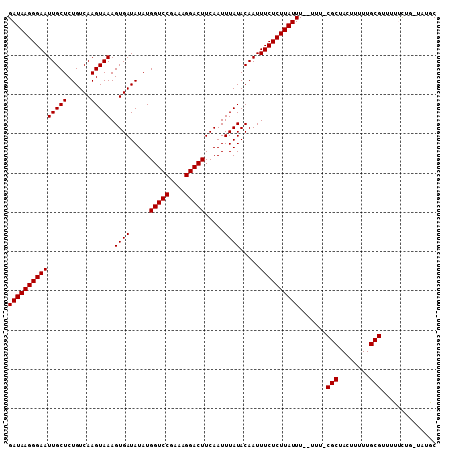
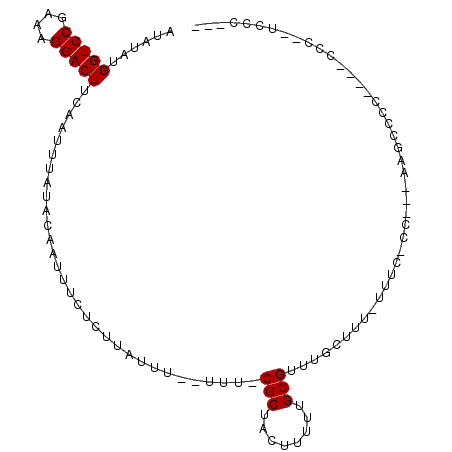
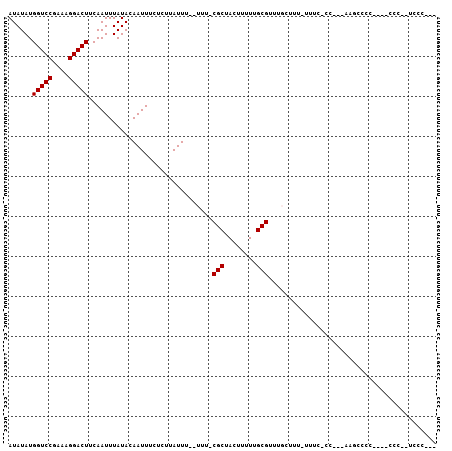
| Location | 6,322,913 – 6,323,004 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -13.83 |
| Consensus MFE | -12.25 |
| Energy contribution | -12.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6322913 91 + 27905053 AUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU--UUU-CGCUACUUUUUGCGUUUGCUUUUUUUC-CC---AAGCCCC----CCA--UCCC--- ......(((((....))))).........................--...-(((........)))...((((......-..---))))...----...--....--- ( -14.00) >DroGri_CAF1 112718 101 + 1 AUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU--UUUCCGCUACUUUUUGCGUUUUUCUG-UAUG-CUUGCUAUCCCUCUCUCUC--UCUCACA .((((((((((....))))).........................--....(((........))).......)-))))-...................--....... ( -13.40) >DroEre_CAF1 94914 86 + 1 AUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU--UUU-CGCUACUUUUUGCGUUUGCUUU--UUC-CC---AAGCCCC----CCUU-------- ......(((((....))))).........................--...-(((........)))...((((.--...-..---))))...----....-------- ( -14.20) >DroWil_CAF1 172716 98 + 1 AUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUUU-CGCUACUUUUUGCGUUUUUCUG-UUUG-CUUUCUCGCUCU----CUC--UCUCACA ......(((((....)))))..............................-(((........)))........-...(-(......))...----...--....... ( -12.90) >DroYak_CAF1 87531 92 + 1 AUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU--UUU-CGCUACUUUUUGCGUUUGCUUU-UUUC-CC---AAGUCCC----CCCCCUCCC--- ......(((((....))))).........................--...-(((........)))........-....-..---.......----.........--- ( -12.30) >DroAna_CAF1 91668 92 + 1 AUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU--UUU-CGCUACUUUUUGCGUUUGCUUU-UUUCGCC---CCGCC------CCU--GCCCGGC ......(((((....))))).........................--...-(((........)))........-....(((---..((.------...--))..))) ( -16.20) >consensus AUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUU__UUU_CGCUACUUUUUGCGUUUGCUUU_UUUC_CC___AAGCCCC____CCC__UCCC___ ......(((((....)))))...............................(((........))).......................................... (-12.25 = -12.25 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:04 2006