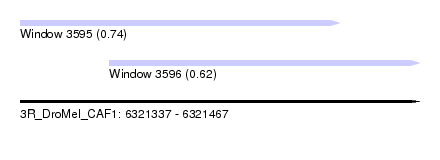
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,321,337 – 6,321,467 |
| Length | 130 |
| Max. P | 0.744951 |

| Location | 6,321,337 – 6,321,441 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -24.81 |
| Energy contribution | -24.75 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6321337 104 + 27905053 UUUGCCAACCCAUUCCGGUCAAAUCUCUAUCACUGCUCACCUGCCAGGUGCCUGGUGACUGCCAACUGGUAACUGGCAACUGGCAACUGGGU--AUCUGGUAAGUG ((((((((((......)))..............((((((..((((((.((((.(....)((((....))))...)))).))))))..)))))--)..))))))).. ( -36.00) >DroSec_CAF1 86169 106 + 1 UUUGCCAACCCAUUCCGGCCAAAUCUCUAUCACUGCUCACCUGCCAGGUGCCUGGUGACUGCCAACUGGUAACGGGUAACUGGCAACUGGGUAUAUCUGGUAAGUG (((((((..((.....))...............((((((..((((((.((((((.....((((....)))).)))))).))))))..))))))....))))))).. ( -34.90) >DroEre_CAF1 93365 97 + 1 UUUGCCAACCCAUUCCGGCCAAAUCUCUGUCACUGCUCACCUGCCAGGUGGCUGGUGACUGCCAACUGGUAACUG-------GUAACUGGGU--AUCUGGUAAGUG ((((((((((((..((((..........((((((((.((((.....)))))).))))))((((....)))).)))-------)....)))))--...))))))).. ( -37.10) >DroYak_CAF1 85922 98 + 1 UUUGCCAACCCAUUCUGGCCAAAUCUCUAUCACUGCUCACCUGCCAGCUGGCUGGUGACUGUUAACUGGUAACUGG------GUAUCUGGGU--AUCUGGUAAGUG ((((((((((((...(((........)))....((((((..((((((.((((.(....).)))).))))))..)))------)))..)))))--...))))))).. ( -31.40) >consensus UUUGCCAACCCAUUCCGGCCAAAUCUCUAUCACUGCUCACCUGCCAGGUGCCUGGUGACUGCCAACUGGUAACUGG______GCAACUGGGU__AUCUGGUAAGUG ((((((((((((.....(((.........((((((..((((.....))))..)))))).((((....))))..........)))...))))).....))))))).. (-24.81 = -24.75 + -0.06)



| Location | 6,321,366 – 6,321,467 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -20.37 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6321366 101 + 27905053 UCACUGCUCACCUGCCAGGUGCCUGGUGACUGCCAACUGGUAACUGGCAACUGGCAACUGGGU--AUCUGGUAAGUGGCGAAUGGUAACGGGCAGAUCACUGC ...((((((...(((((((((((..((...(((((..((.(....).))..)))))))..)))--)))))))).((.((.....)).))))))))........ ( -40.80) >DroSec_CAF1 86198 103 + 1 UCACUGCUCACCUGCCAGGUGCCUGGUGACUGCCAACUGGUAACGGGUAACUGGCAACUGGGUAUAUCUGGUAAGUGGCGAAUGGUAACGGGCAGAUCACUGC ...((((((...(((((((((((..((...(((((.(((....))).....)))))))..)))..)))))))).((.((.....)).))))))))........ ( -37.20) >DroEre_CAF1 93394 85 + 1 UCACUGCUCACCUGCCAGGUGGCUGGUGACUGCCAACUGGUAACUG-------GUAACUGGGU--AUCUGGUAAGUGGCGAAUGGUAA---------CACUGC ....(((.(((.(((((((((.(..((.((((((....)))....)-------)).))..).)--)))))))).))))))........---------...... ( -30.20) >DroYak_CAF1 85951 81 + 1 UCACUGCUCACCUGCCAGCUGGCUGGUGACUGUUAACUGGUAACUGG------GUAUCUGGGU--AUCUGGUAAGUGGCAGAU--------------CACUGC ...((((.(((.((((((.((((.(....).)))).))))))((..(------((((....))--)))..))..)))))))..--------------...... ( -31.70) >consensus UCACUGCUCACCUGCCAGGUGCCUGGUGACUGCCAACUGGUAACUGG______GCAACUGGGU__AUCUGGUAAGUGGCGAAUGGUAA_________CACUGC ....(((.(((.(((((((((((..((...((((....))))..............))..)))..)))))))).))))))....................... (-20.37 = -20.80 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:02 2006