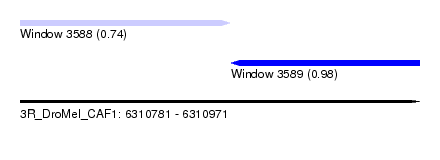
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,310,781 – 6,310,971 |
| Length | 190 |
| Max. P | 0.980654 |

| Location | 6,310,781 – 6,310,881 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -13.64 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6310781 100 + 27905053 GUAAAAACACAA--CUCCGGCA---ACUUGACGACGGCAGCG--CAGACUGCGCCAAAAGUCAAAUUUUGUAGAUUGCAAUAAAUCAAAUAGCAAUUACACGAAAUU ............--....(...---.)(((((...(((.(((--.....))))))....)))))(((((((.((((((.............))))))..))))))). ( -19.42) >DroPse_CAF1 73172 99 + 1 GCAAGUUGACGACUCUCUGGGA---CUUUGU---CGGCGGCA--CAGACUGCGCCAAAAGUCAAAUUUUGUAGAUUGCAAUAAAUCAAAUAGCAAUUACACGAAAUU ((..((((((((.((.....))---..))))---)))).)).--..((((........)))).((((((((.((((((.............))))))..)))))))) ( -22.72) >DroEre_CAF1 83253 84 + 1 -----AA-------------CA---ACUUGACGACGGCGGCG--CAGACUGCGCCAAAAGUCAAAUUUUGUAGAUUGCAAUAAAUCAAAUAGCAAUUACACGAAAUU -----..-------------..---.......(((...((((--(.....)))))....))).((((((((.((((((.............))))))..)))))))) ( -22.02) >DroYak_CAF1 74821 89 + 1 GUAAAAA-------------CA---ACUUGACGACGGCAGCG--CAGACUGCGCCAAAAGUCAAAUUUUCUAGAUUGCAAUAAAUCAAAUAGCAAUUACACGAAAUU ((((...-------------..---..(((((...(((.(((--.....))))))....)))))........((((......)))).........))))........ ( -15.50) >DroAna_CAF1 79060 103 + 1 --ACACAGGCCG--CUCCGGCAGGCAGUUGAUGGAGGCAGCGCCCAGACUGCGCCAAAAGUCAAAUUUUCCAGAUUGCAAUAAAUCAAAUGGCAAUUACACGAAAUU --......((((--(....))..((((((..(((((((...)))..((((........))))......))))))))))............))).............. ( -23.90) >DroPer_CAF1 78429 99 + 1 GCAAGUUGACGACUCUCUGGGA---CUUUGU---CGGCGGCA--CAGACUGCGCCAAAAGUCAAAUUUUGUAGAUUGCUAUAAAUCAAAUAGCAAUUACACGAAAUU ((..((((((((.((.....))---..))))---)))).)).--..((((........)))).((((((((.(((((((((.......)))))))))..)))))))) ( -27.10) >consensus G_AAAAAGAC_A__CUC_GGCA___ACUUGACGACGGCAGCG__CAGACUGCGCCAAAAGUCAAAUUUUGUAGAUUGCAAUAAAUCAAAUAGCAAUUACACGAAAUU ...........................(((((...(((.(((.......))))))....)))))(((((((.((((((.............))))))..))))))). (-13.64 = -14.50 + 0.86)
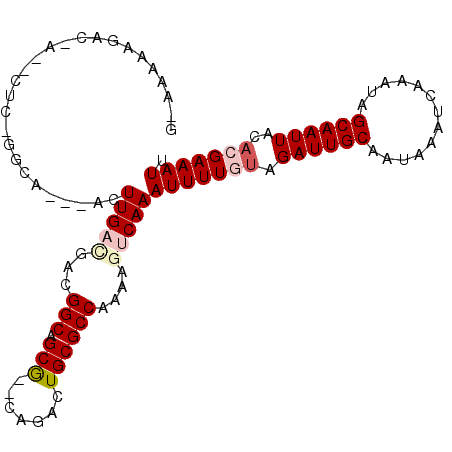
| Location | 6,310,881 – 6,310,971 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 79.95 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -10.34 |
| Energy contribution | -12.16 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.24 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6310881 90 - 27905053 CCCACACACAUGCACACACACACACUAGCUCCUUUCAAUCCCCCGCUUAGCAAUGUGUGUG--UAUGUGUGCAAAAUUGAGAAAACAUGCCU ....((((((((((((((((......(((...............)))......))))))))--))))))))..................... ( -27.76) >DroSec_CAF1 75751 82 - 1 CCCACACACA----CAUACACACACUAGCUCCUUUCAAUCCCCCGCUUUGCAAUGUGUG------UGUGUGCAAAAUUGAGAAAACAUGCCU ....((((((----((((((......(((...............)))......))))))------))))))..................... ( -19.16) >DroSim_CAF1 81558 88 - 1 CCCACACACA----CGUACACACACUAGCUCCUUUCAAUCCCCCGCUUUGCCAUGUGUGUGUGUGUGUGUGCAAAAUUGAGAAAACAUGCCU ..((((((((----((((((((((...((....................))..))))))))))))))))))..................... ( -27.05) >DroEre_CAF1 83337 82 - 1 CCCACACACA------CACACACACUAGCUCCUUUCAAUCCCCCGCCUAGCCA--UGUGUG--UGUGUGUGGAAAAUUGAGAAAACAUGCCU .(((((((((------((((((..(((((...............).))))...--))))))--))))))))).................... ( -29.16) >DroYak_CAF1 74910 84 - 1 CCCACACACA------UACACACACUAGCUCCUUUCAAUCCCCCGCCUAACAAAGUGUGUG--UGUGUGUGCAAAAUUGAGAAAACAUGCCU ....((((((------((((((((((...........................))))))))--))))))))..................... ( -24.13) >DroAna_CAF1 79163 68 - 1 CCCACACACG-------------------UCCUCCCAG-CCACCCCCUGGCCG--GGGCUG--UGUGUGUGCGAAAUUGAGAAAACAUGCCU .(((((((((-------------------...((((.(-(((.....)))).)--)))...--)))))))).)................... ( -22.90) >consensus CCCACACACA______UACACACACUAGCUCCUUUCAAUCCCCCGCCUAGCAAUGUGUGUG__UGUGUGUGCAAAAUUGAGAAAACAUGCCU ..((((((((......(((((((...............................)))))))..))))))))..................... (-10.34 = -12.16 + 1.82)

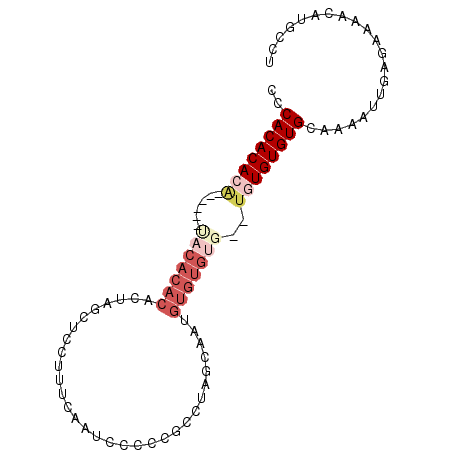
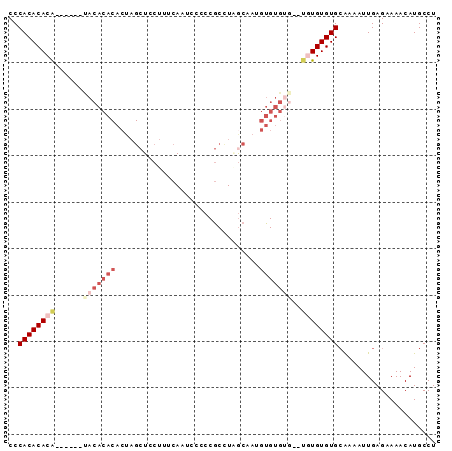
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:55 2006