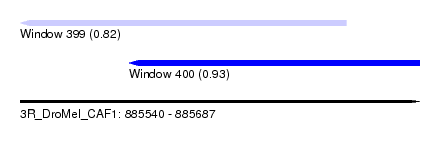
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 885,540 – 885,687 |
| Length | 147 |
| Max. P | 0.932209 |

| Location | 885,540 – 885,660 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -26.75 |
| Energy contribution | -27.53 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 885540 120 - 27905053 AUUGAAGGCGGCCGAAGGUUGUCGACUGUCCGACUGCUCGCUCGGCUGCUAUCAAUGAGUGCGCUGUCAAAAUCAAAAUGCUGUGCAUUUAUGGGUACAAAUCCACGAAAUGGAAAGUAG ..(((.(((((((((.(((.((((......)))).)).)..))))))))).)))((((((((((.((............)).))))))))))...(((...((((.....))))..))). ( -40.40) >DroSec_CAF1 53421 105 - 1 AUUGAAGGCGGCCGAAGGUUGUCGACUG------------CUCGGCUGCUGGCAAUGAGUGCUGUGCC---AUCGAAAUGCCUUGCAUUUAUGGGUACAAAUCCACGAAAUGGAAAGUAG ((((..(((((((((.((((...)))).------------.)))))))))..))))...(((((((((---....((((((...))))))...)))))...((((.....)))).)))). ( -39.50) >DroSim_CAF1 52917 105 - 1 AUUGAAGGCGGCCGAAGGUUGUCGACUG------------CUCGGCUGCUGUCAAUGGGUGCUAUGCC---AUCGAAAUGCCUUGCAUUUCUGGGUACAAAUCCACGAAAUGGAAAGUAG (((((.(((((((((.((((...)))).------------.))))))))).))))).(((.....)))---.......(((.((.(((((((((((....))))).)))))).)).))). ( -39.20) >consensus AUUGAAGGCGGCCGAAGGUUGUCGACUG____________CUCGGCUGCUGUCAAUGAGUGCUAUGCC___AUCGAAAUGCCUUGCAUUUAUGGGUACAAAUCCACGAAAUGGAAAGUAG (((((.(((((((((.((((...))))..............))))))))).)))))...((((...((....(((((((((...)))))).(((((....))))))))...))..)))). (-26.75 = -27.53 + 0.78)

| Location | 885,580 – 885,687 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -24.15 |
| Energy contribution | -24.26 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 885580 107 - 27905053 -------------GCUGAUUGAUUCUAAUUGAAAGCGGCAAUUGAAGGCGGCCGAAGGUUGUCGACUGUCCGACUGCUCGCUCGGCUGCUAUCAAUGAGUGCGCUGUCAAAAUCAAAAUG -------------.....((((((....((((.((((((.(((((.(((((((((.(((.((((......)))).)).)..))))))))).)))))..)).)))).)))))))))).... ( -41.30) >DroSec_CAF1 53461 105 - 1 UGUGCAGUUGACAGCUGAUUGAUUCUAAUUGAAAGCGGCAAUUGAAGGCGGCCGAAGGUUGUCGACUG------------CUCGGCUGCUGGCAAUGAGUGCUGUGCC---AUCGAAAUG .(.(((((((((((((..(((.(((.....))).((.((........)).))))).))))))))))))------------).)(((.((.((((.....)))))))))---......... ( -37.40) >DroSim_CAF1 52957 105 - 1 UGUGCAGUUGACAGCUGAUUGAUUCUAAUUGAAAGCGGCAAUUGAAGGCGGCCGAAGGUUGUCGACUG------------CUCGGCUGCUGUCAAUGGGUGCUAUGCC---AUCGAAAUG .(.(((((((((((((..(((.(((.....))).((.((........)).))))).))))))))))))------------).)(((.((...........))...)))---......... ( -34.90) >consensus UGUGCAGUUGACAGCUGAUUGAUUCUAAUUGAAAGCGGCAAUUGAAGGCGGCCGAAGGUUGUCGACUG____________CUCGGCUGCUGUCAAUGAGUGCUAUGCC___AUCGAAAUG .............(((..(..((....))..).)))(((((((((.(((((((((.((((...))))..............))))))))).)))))........))))............ (-24.15 = -24.26 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:02 2006