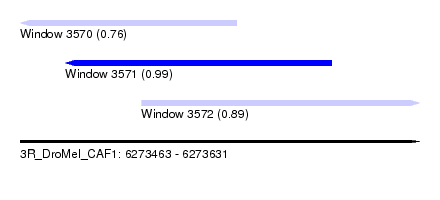
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,273,463 – 6,273,631 |
| Length | 168 |
| Max. P | 0.994739 |

| Location | 6,273,463 – 6,273,554 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.23 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
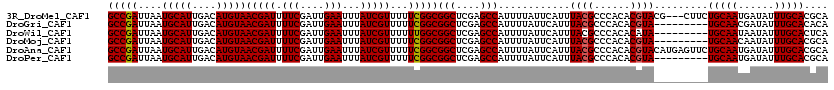
>3R_DroMel_CAF1 6273463 91 - 27905053 AUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CC-CU------UGC------UGUCCC-----UGUCCCC-CAU ..............(((((.((((....))))))).))((((((((...))))))))((((((.....--.)-))------)))------......-----.......-... ( -19.70) >DroVir_CAF1 40632 96 - 1 AUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CC-G--GC-ACUCCUCUGAAUGUCUC---------UCC-CAC ...................((((((..(((........((((((((...)))))))))))..))))))--..-(--((-(.((....)).))))..---------...-... ( -19.20) >DroWil_CAF1 50443 103 - 1 AUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CCACCAGCAUGUCC------CCUCUCUCAAUUGUCCAC-CAU ...((((.((....(((((.((((....))))))).))((((((((...))))))))....(((((((--(.....))))))))------......)).)))).....-... ( -22.90) >DroYak_CAF1 40508 92 - 1 AUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CCCCU------UGC------UGUCCC-----UGUCUCC-CUU ..............(((((.((((....))))))).))((((((((...))))))))((((((.....--..)))------)))------......-----.......-... ( -19.20) >DroAna_CAF1 44793 86 - 1 AUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUGUGUGUC-CU------CUC------CGUCCC-----------A-CA- .................((((((((..(((........((((((((...)))))))))))..))))))))..-..------...------......-----------.-..- ( -17.30) >DroPer_CAF1 40396 98 - 1 AUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CC-CCGCCAAGUCC------UCUCCC-----UCUCCUGCCCU ........((....((...((((((..(((........((((((((...)))))))))))..))))))--..-.)).....)).------......-----........... ( -16.20) >consensus AUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG__CC_CU______UCC______UGUCCC_____UGUCCCC_CAU ...................((((((..(((........((((((((...)))))))))))..))))))............................................ (-15.37 = -15.23 + -0.14)

| Location | 6,273,482 – 6,273,594 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -24.57 |
| Energy contribution | -24.43 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6273482 112 - 27905053 GUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CC-CU----- ...(((((.......(((....))).........((((.......))))....))))).((((((..(((........((((((((...)))))))))))..))))))--..-..----- ( -24.90) >DroPse_CAF1 35118 117 - 1 GUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CC-CCGCCAA ..............(((((((......)))..((((((...(((....))).)))))).((((((..(((........((((((((...)))))))))))..))))))--..-..)))). ( -27.50) >DroWil_CAF1 50468 118 - 1 GUAAAUGAAUAAAAUGGCUCGAGCCGCCAAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CCACCAGCAU ...............(((....)))((.....((((((...(((....))).)))))).((((((..(((........((((((((...)))))))))))..))))))--......)).. ( -26.60) >DroMoj_CAF1 60520 115 - 1 GUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CC-G--AAAA ...............(((....)))..((...((((((...(((....))).)))))).((((((..(((........((((((((...)))))))))))..))))))--.)-)--.... ( -25.40) >DroAna_CAF1 44805 114 - 1 GUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUGUGUGUC-CU----- ...............(((....)))(((((..((((((...(((....))).))))))..((((....))))...)))((((((((...)))))))))).(((((....)))-))----- ( -26.20) >DroPer_CAF1 40417 117 - 1 GUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG--CC-CCGCCAA ..............(((((((......)))..((((((...(((....))).)))))).((((((..(((........((((((((...)))))))))))..))))))--..-..)))). ( -27.50) >consensus GUAAAUGAAUAAAAUGGCUCGAGCCGCCGAAAAACGAUAAAUUCAAUCGAAAAUCGUUACAUGUCAAUGCAUUAAUCGGCCAUAAUCCCAUUAUGGCGCAAGGAUAUG__CC_CC__CAA ...(((((.......(((....))).........((((.......))))....))))).((((((..(((........((((((((...)))))))))))..))))))............ (-24.57 = -24.43 + -0.14)

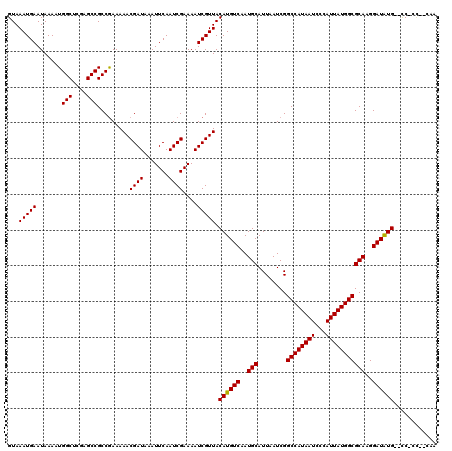
| Location | 6,273,514 – 6,273,631 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.23 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -23.85 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
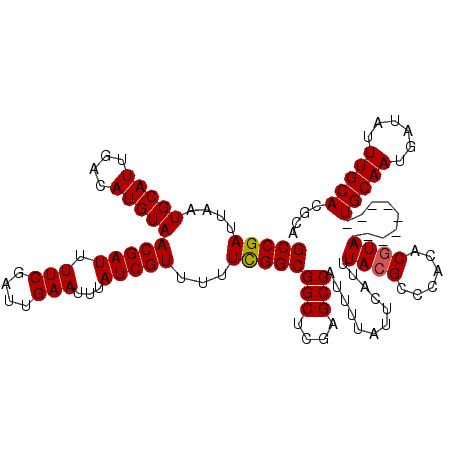
>3R_DroMel_CAF1 6273514 117 + 27905053 GCCGAUUAAUGCAUUGACAUGUAACGAUUUUCGAUUGAAUUUAUCGUUUUUCGGCGGCUCGAGCCAUUUUAUUCAUUUACGCCCACACGUACG---CUUCUGCAAUGAUAUUUGCACGCA (((((....(((((....)))))(((((.(((....)))...)))))...)))))(((....)))............((((......)))).(---(...(((((......))))).)). ( -27.60) >DroGri_CAF1 55341 111 + 1 GCCGAUUAAUGCAUUGACAUGUAACGAUUUUCGAUUGAAUUUAUCGUUUUUCGGCGGCUCGAGCCAUUUUAUUCAUUUACGCCCACACGUA---------UGCAACGAUAUUUGCACACA (((((....(((((....)))))(((((.(((....)))...)))))...)))))(((....))).......................((.---------(((((......))))).)). ( -25.40) >DroWil_CAF1 50506 111 + 1 GCCGAUUAAUGCAUUGACAUGUAACGAUUUUCGAUUGAAUUUAUCGUUUUUUGGCGGCUCGAGCCAUUUUAUUCAUUUACGCCCACACAUA---------UGCAAUAAUAUUUGCACUCA (((((....(((((....)))))(((((.(((....)))...)))))...)))))(((....)))..........................---------(((((......))))).... ( -22.00) >DroMoj_CAF1 60555 111 + 1 GCCGAUUAAUGCAUUGACAUGUAACGAUUUUCGAUUGAAUUUAUCGUUUUUCGGCGGCUCGAGCCAUUUUAUUCAUUUACGCCCACACGUA---------UGCAACAAUAUUUGCACGCA (((((....(((((....)))))(((((.(((....)))...)))))...)))))(((....))).......................(..---------(((((......)))))..). ( -26.20) >DroAna_CAF1 44839 120 + 1 GCCGAUUAAUGCAUUGACAUGUAACGAUUUUCGAUUGAAUUUAUCGUUUUUCGGCGGCUCGAGCCAUUUUAUUCAUUUACGCCCACACGUACAUGAGUUCUGCAAUGAUAUUUGCACGCA (((((....(((((....)))))(((((.(((....)))...)))))...)))))(((....))).....((((((.((((......)))).))))))..(((((......))))).... ( -31.20) >DroPer_CAF1 40454 111 + 1 GCCGAUUAAUGCAUUGACAUGUAACGAUUUUCGAUUGAAUUUAUCGUUUUUCGGCGGCUCGAGCCAUUUUAUUCAUUUACGCCCACACGUA---------UGCAAUGAUAUUUGCACGCA (((((....(((((....)))))(((((.(((....)))...)))))...)))))(((....))).......................(..---------(((((......)))))..). ( -27.10) >consensus GCCGAUUAAUGCAUUGACAUGUAACGAUUUUCGAUUGAAUUUAUCGUUUUUCGGCGGCUCGAGCCAUUUUAUUCAUUUACGCCCACACGUA_________UGCAAUGAUAUUUGCACGCA (((((....(((((....)))))(((((.(((....)))...)))))...)))))(((....)))............((((......)))).........(((((......))))).... (-23.85 = -23.88 + 0.03)
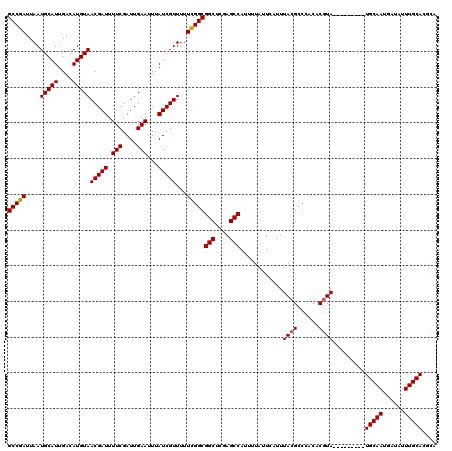
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:39 2006