
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,271,198 – 6,271,294 |
| Length | 96 |
| Max. P | 0.887022 |

| Location | 6,271,198 – 6,271,294 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -20.17 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6271198 96 + 27905053 GCUUUUCCUGCUUUUCCCGCUUUAGCAGCC-UUUGCUGCCUUUGCUGCUCGUGCUGAUAACGUGG----CCAGCAGCGUUGGUCAGCAGUGGUCGGAUAAA .....(((((((...........))))(((-.((((((((..(((((((.(.(((........))----)))))))))..)).)))))).))).))).... ( -35.70) >DroSec_CAF1 36421 96 + 1 GCUUUUCCUGCUUUUCCCGCUUUCGCAGCC-UUUGCUGCCUUUGCUGCUCGUGCUGAUAACGUGG----CCAGCAGCGUUGGUCAGCAGUGGUCGGAUAAA ((.......))...(((.((....)).(((-.((((((((..(((((((.(.(((........))----)))))))))..)).)))))).))).))).... ( -35.00) >DroSim_CAF1 39507 96 + 1 GCUUUUCCUGCUUUUCCCGCUUUCGCAGCC-UUUGCUGCCUUUGCUGCUCGUGCUGAUAACGUGG----CCAGCAGCGUUGGUCAGCAGUGGUCGGAUAAA ((.......))...(((.((....)).(((-.((((((((..(((((((.(.(((........))----)))))))))..)).)))))).))).))).... ( -35.00) >DroAna_CAF1 42794 95 + 1 GAUUUUGCUUCU------GCUUUCGCAAUAGUAUCCUGGCGCUGCCUCGCCGGCUGAUAACGGGGCCGGCCAACAGCGCUGGUCAGCAGAGGCCGGAUAAA .(((..((((((------(((...((....))...(..((((((....(((((((........)))))))...))))))..)..)))))))))..)))... ( -43.50) >consensus GCUUUUCCUGCUUUUCCCGCUUUCGCAGCC_UUUGCUGCCUUUGCUGCUCGUGCUGAUAACGUGG____CCAGCAGCGUUGGUCAGCAGUGGUCGGAUAAA ((.......)).....(((.....(((((.....)))))....((((((...(((((((((((((....))....))))).)))))))))))))))..... (-20.17 = -21.05 + 0.88)


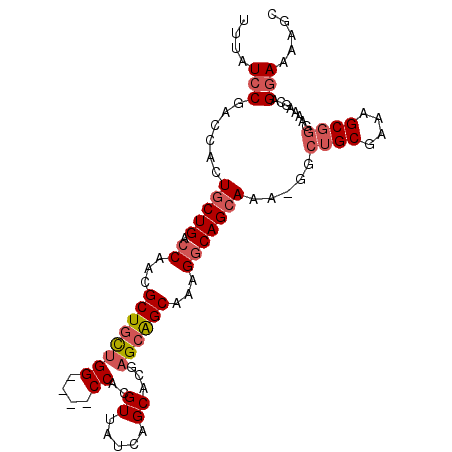
| Location | 6,271,198 – 6,271,294 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -20.42 |
| Energy contribution | -21.80 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.887022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6271198 96 - 27905053 UUUAUCCGACCACUGCUGACCAACGCUGCUGG----CCACGUUAUCAGCACGAGCAGCAAAGGCAGCAAA-GGCUGCUAAAGCGGGAAAAGCAGGAAAAGC ............(((((..((..(((((((((----(..........)).).))))))...((((((...-.))))))...)..))...)))))....... ( -31.10) >DroSec_CAF1 36421 96 - 1 UUUAUCCGACCACUGCUGACCAACGCUGCUGG----CCACGUUAUCAGCACGAGCAGCAAAGGCAGCAAA-GGCUGCGAAAGCGGGAAAAGCAGGAAAAGC ....(((..((..(((((.((...((((((((----(..........)).).))))))...)))))))..-))((((....))))........)))..... ( -33.30) >DroSim_CAF1 39507 96 - 1 UUUAUCCGACCACUGCUGACCAACGCUGCUGG----CCACGUUAUCAGCACGAGCAGCAAAGGCAGCAAA-GGCUGCGAAAGCGGGAAAAGCAGGAAAAGC ....(((..((..(((((.((...((((((((----(..........)).).))))))...)))))))..-))((((....))))........)))..... ( -33.30) >DroAna_CAF1 42794 95 - 1 UUUAUCCGGCCUCUGCUGACCAGCGCUGUUGGCCGGCCCCGUUAUCAGCCGGCGAGGCAGCGCCAGGAUACUAUUGCGAAAGC------AGAAGCAAAAUC ..((((((((....(((....)))((((((.((((((..........))))))..))))))))).)))))((.((((....))------)).))....... ( -41.20) >consensus UUUAUCCGACCACUGCUGACCAACGCUGCUGG____CCACGUUAUCAGCACGAGCAGCAAAGGCAGCAAA_GGCUGCGAAAGCGGGAAAAGCAGGAAAAGC ....(((......(((((.((...((((((((....))..((.....))...))))))...))))))).....((((....))))........)))..... (-20.42 = -21.80 + 1.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:36 2006