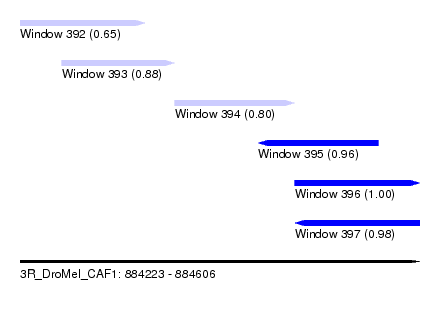
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 884,223 – 884,606 |
| Length | 383 |
| Max. P | 0.995138 |

| Location | 884,223 – 884,343 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.10 |
| Mean single sequence MFE | -33.43 |
| Consensus MFE | -30.07 |
| Energy contribution | -30.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 884223 120 + 27905053 UAUCGUAAAACAAAGUGUAUAAUUUUACAUUUAAAUGCGCUUAAGCCAGCUUUUAAUGAAAUGUAAGCUUUUGAUUGGGGGCUGAGCUGGGGCUUGGGGUGGGAUCGUAUCGGAUUCCGA .(((.(((....(((((((......)))))))......(((..((((((((((((((.(((........))).))))))))))).)))..)))))).)))((((((......)))))).. ( -37.40) >DroSec_CAF1 52068 119 + 1 UAUCGUAAAACAAAUUGUAUAAUUUUACAUUUAAAUGCGCUUAAGCCAGCUCUUAAUGAAAUGUAAGAUUUUGAUUUG-GGCUGAGCUGGGGCUUGGGGUGGGAUCGUGUCGGAUUCCGA .(((.(((.......((((......)))).........(((..(((((((((..(((.(((........))).))).)-))))).)))..)))))).)))((((((......)))))).. ( -31.30) >DroSim_CAF1 51551 119 + 1 UAUCGUAAAACAAAUUGUAUAAUUUUACAUUUAAAUGCGCUUAAGCCAGCUCUUAAUAAAAUGUAAGCUUUUGAUUGG-GGCUGAGCUGGGGCUUGGGGUGGGAUCGUAUCGGAUUCCGA .(((.(((.......((((......)))).........(((..((((((((((...(((((.(....))))))...))-))))).)))..)))))).)))((((((......)))))).. ( -31.60) >consensus UAUCGUAAAACAAAUUGUAUAAUUUUACAUUUAAAUGCGCUUAAGCCAGCUCUUAAUGAAAUGUAAGCUUUUGAUUGG_GGCUGAGCUGGGGCUUGGGGUGGGAUCGUAUCGGAUUCCGA .(((.(((.......((((......)))).........(((..((((((((.(((((.(((........))).))))).))))).)))..)))))).)))((((((......)))))).. (-30.07 = -30.40 + 0.33)

| Location | 884,263 – 884,371 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.69 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -32.35 |
| Energy contribution | -32.46 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 884263 108 + 27905053 UUAAGCCAGCUUUUAAUGAAAUGUAAGCUUUUGAUUGGGGGCUGAGCUGGGGCUUGGGGUGGGAUCGUAUCGGAUUCCGAUU------------CCGAUUUCGAUUCGGGUUCGGAAUUC ...((((((((((((((.(((........))).))))))))))).))).(((((((((.((..((((.(((((...))))).------------.))))..)).)))))))))....... ( -39.70) >DroSec_CAF1 52108 119 + 1 UUAAGCCAGCUCUUAAUGAAAUGUAAGAUUUUGAUUUG-GGCUGAGCUGGGGCUUGGGGUGGGAUCGUGUCGGAUUCCGAUUCCAAUUCCAAUUCCGAUUUCGAUUCGGGUUCGGAAUUC ...(((((((((..(((.(((........))).))).)-))))).))).((..(((((((((((((......)))))).)))))))..))((((((((.((((...)))).)))))))). ( -43.40) >DroSim_CAF1 51591 119 + 1 UUAAGCCAGCUCUUAAUAAAAUGUAAGCUUUUGAUUGG-GGCUGAGCUGGGGCUUGGGGUGGGAUCGUAUCGGAUUCCGAUUCCGAUUCCAAUUCCGAUUUCGAUUCGGGUUCGGAAUUC ...((((((((((...(((((.(....))))))...))-))))).))).((..(((((((((((((......)))))).)))))))..))((((((((.((((...)))).)))))))). ( -43.40) >consensus UUAAGCCAGCUCUUAAUGAAAUGUAAGCUUUUGAUUGG_GGCUGAGCUGGGGCUUGGGGUGGGAUCGUAUCGGAUUCCGAUUCC_AUUCCAAUUCCGAUUUCGAUUCGGGUUCGGAAUUC ...((((((((.(((((.(((........))).))))).))))).))).(((((((((.((..((((.(((((...)))))..............))))..)).)))))))))....... (-32.35 = -32.46 + 0.11)

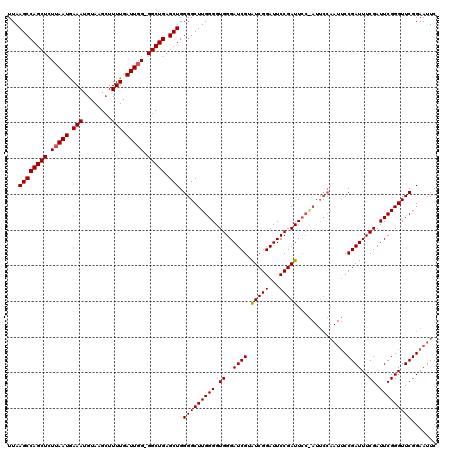
| Location | 884,371 – 884,486 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -24.91 |
| Consensus MFE | -23.93 |
| Energy contribution | -24.15 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 884371 115 + 27905053 GGUUUCACCUUCGCCAUGCGUGCGAGACCUCCAAAAAUGCGGCAAGGCAAAAAUCGAACGCAAAUGAUUUAUAAUUAAAUAUUUUGUGAUUAUUAAGAA-AAACGAA----UAAAAAAAC (((((..((((.(((..((((..((....)).....)))))))))))...)))))...((((((..(((((....)))))..))))))...........-.......----......... ( -19.50) >DroSec_CAF1 52227 119 + 1 GGUUUCACCUUCGCCAUGCGUGCGAGACCUCCAAAAACGCGGCAAGGCGAAAAUCGAACGCAAAUGAUUUAUAAUUAAAUAUUUUGUGAUUAUCAAGAAGAAACGAAGGAAAA-AAAAAC .(((((...((((((.(((.((((.............))))))).)))))).......((((((..(((((....)))))..))))))...........))))).........-...... ( -29.12) >DroSim_CAF1 51710 120 + 1 GGUUUCACCUUCGCCAUGCGUGCGAGACCUCCAAAAAUGCGGCGAGGCGAAAAUCGAACGCAAAUGAUUUAUAAUUAAAUAUUUUGUGAUUAUCAAGAAGAAACGAAGGAAAAAAAAAAC .(((((...((((((.(((.((((.............))))))).)))))).......((((((..(((((....)))))..))))))...........)))))................ ( -26.12) >consensus GGUUUCACCUUCGCCAUGCGUGCGAGACCUCCAAAAAUGCGGCAAGGCGAAAAUCGAACGCAAAUGAUUUAUAAUUAAAUAUUUUGUGAUUAUCAAGAAGAAACGAAGGAAAAAAAAAAC .(((((...((((((.(((.((((.............))))))).)))))).......((((((..(((((....)))))..))))))...........)))))................ (-23.93 = -24.15 + 0.23)

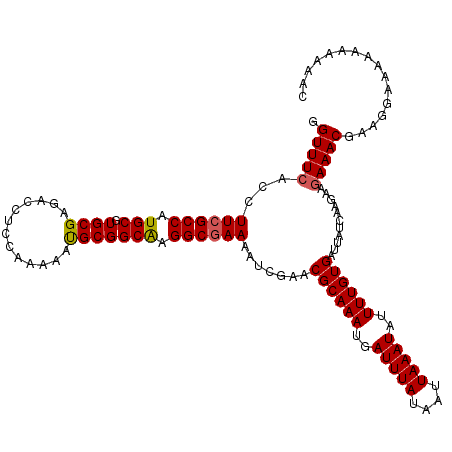
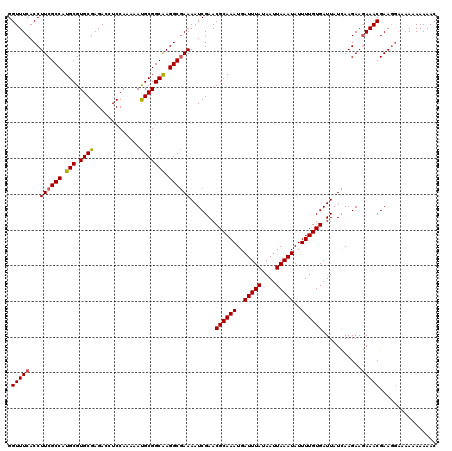
| Location | 884,451 – 884,566 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 884451 115 - 27905053 UAUGCUGAGCAUAAAUUUUCUGCAACGUCGCUUAAUGCGAUUUUGUUUCGUAUAUUUUCUGCUAAUUGCUUGGCGCUUUUGUUUUUUUA----UUCGUUU-UUCUUAAUAAUCACAAAAU ...((..((((..........((((.(((((.....))))).))))...(((.......)))....))))..))..((((((....(((----((.....-.....)))))..)))))). ( -22.10) >DroSec_CAF1 52307 119 - 1 UAUGCUAAGCAUAAAUUUUCUGCAACGUCGCUUAAUGCGAUUUUGUUUCGUAUAUUUUCUGCUAAUUGCUUGGCGCUUUUGUUUUU-UUUUCCUUCGUUUCUUCUUGAUAAUCACAAAAU ...((((((((..........((((.(((((.....))))).))))...(((.......)))....))))))))..((((((..((-.......(((........))).))..)))))). ( -21.80) >DroSim_CAF1 51790 120 - 1 UAUGCUAAGCAUAAAUUUUCUGCAACGUCGCUUAAUGCGAUUUUGUUUCGUAUAUUUUCUGCUAAUUGCUUGGCGCUUUUGUUUUUUUUUUCCUUCGUUUCUUCUUGAUAAUCACAAAAU ...((((((((..........((((.(((((.....))))).))))...(((.......)))....))))))))..((((((..((........(((........))).))..)))))). ( -21.70) >consensus UAUGCUAAGCAUAAAUUUUCUGCAACGUCGCUUAAUGCGAUUUUGUUUCGUAUAUUUUCUGCUAAUUGCUUGGCGCUUUUGUUUUUUUUUUCCUUCGUUUCUUCUUGAUAAUCACAAAAU ...((((((((..........((((.(((((.....))))).))))...(((.......)))....)))))))).............................................. (-21.32 = -21.10 + -0.22)

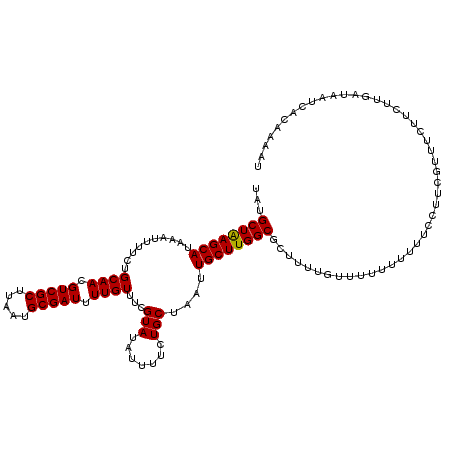
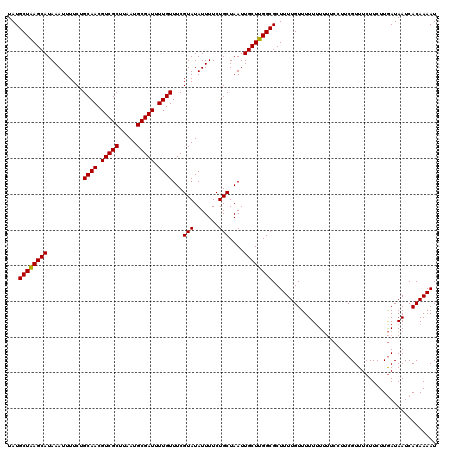
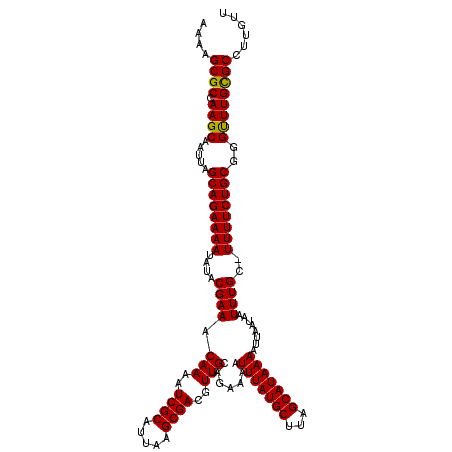
| Location | 884,486 – 884,606 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -31.97 |
| Energy contribution | -31.53 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.16 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 884486 120 + 27905053 AAAAGCGCCAAGCAAUUAGCAGAAAAUAUACGAAACAAAAUCGCAUUAAGCGACGUUGCAGAAAAUUUAUGCUCAGCAUAAAAUUAAUAAUUUGCCUUUUCUGCGGGCUUGUGCCUUGUU ..(((.(((((((.....((((((((....((((.(((..((((.....))))..))).......(((((((...)))))))........))))..))))))))..))))).)))))... ( -32.80) >DroSec_CAF1 52346 119 + 1 AAAAGCGCCAAGCAAUUAGCAGAAAAUAUACGAAACAAAAUCGCAUUAAGCGACGUUGCAGAAAAUUUAUGCUUAGCAUAAAAUUAAUAAUUUGC-UUUUCUGCGGGUUUGCGCCUUGUU ....((((.((((.....((((((((....((((.(((..((((.....))))..))).......(((((((...)))))))........)))).-))))))))..))))))))...... ( -30.60) >DroSim_CAF1 51830 119 + 1 AAAAGCGCCAAGCAAUUAGCAGAAAAUAUACGAAACAAAAUCGCAUUAAGCGACGUUGCAGAAAAUUUAUGCUUAGCAUAAAAUUAAUAAUUUGC-UUUUCUGCGGGUUUGCGCCUUGUU ....((((.((((.....((((((((....((((.(((..((((.....))))..))).......(((((((...)))))))........)))).-))))))))..))))))))...... ( -30.60) >consensus AAAAGCGCCAAGCAAUUAGCAGAAAAUAUACGAAACAAAAUCGCAUUAAGCGACGUUGCAGAAAAUUUAUGCUUAGCAUAAAAUUAAUAAUUUGC_UUUUCUGCGGGUUUGCGCCUUGUU ....((((.((((.....((((((((....((((.(((..((((.....))))..))).......(((((((...)))))))........))))..))))))))..))))))))...... (-31.97 = -31.53 + -0.44)

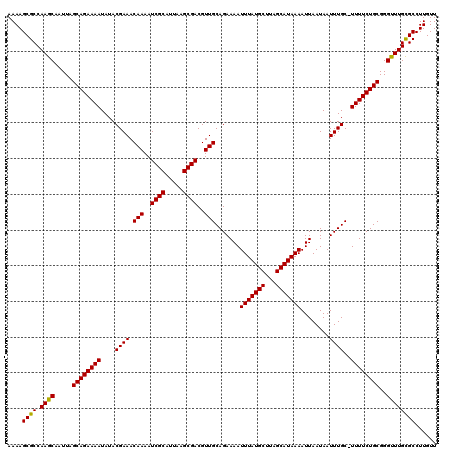
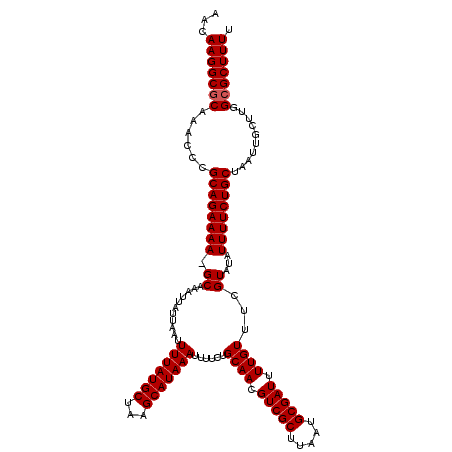
| Location | 884,486 – 884,606 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -30.59 |
| Energy contribution | -30.92 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 884486 120 - 27905053 AACAAGGCACAAGCCCGCAGAAAAGGCAAAUUAUUAAUUUUAUGCUGAGCAUAAAUUUUCUGCAACGUCGCUUAAUGCGAUUUUGUUUCGUAUAUUUUCUGCUAAUUGCUUGGCGCUUUU ...(((((.(((((..((((((((.((...........(((((((...)))))))......((((.(((((.....))))).))))...))...)))))))).....)))))..))))). ( -35.70) >DroSec_CAF1 52346 119 - 1 AACAAGGCGCAAACCCGCAGAAAA-GCAAAUUAUUAAUUUUAUGCUAAGCAUAAAUUUUCUGCAACGUCGCUUAAUGCGAUUUUGUUUCGUAUAUUUUCUGCUAAUUGCUUGGCGCUUUU ...(((((((......((((((((-.............(((((((...)))))))......((((.(((((.....))))).))))........))))))))..........))))))). ( -31.39) >DroSim_CAF1 51830 119 - 1 AACAAGGCGCAAACCCGCAGAAAA-GCAAAUUAUUAAUUUUAUGCUAAGCAUAAAUUUUCUGCAACGUCGCUUAAUGCGAUUUUGUUUCGUAUAUUUUCUGCUAAUUGCUUGGCGCUUUU ...(((((((......((((((((-.............(((((((...)))))))......((((.(((((.....))))).))))........))))))))..........))))))). ( -31.39) >consensus AACAAGGCGCAAACCCGCAGAAAA_GCAAAUUAUUAAUUUUAUGCUAAGCAUAAAUUUUCUGCAACGUCGCUUAAUGCGAUUUUGUUUCGUAUAUUUUCUGCUAAUUGCUUGGCGCUUUU ...(((((((......((((((((.((...........(((((((...)))))))......((((.(((((.....))))).))))...))...))))))))..........))))))). (-30.59 = -30.92 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:59 2006