| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,259,965 – 6,260,057 |
| Length | 92 |
| Max. P | 0.984297 |

| Location | 6,259,965 – 6,260,057 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 73.20 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -10.24 |
| Energy contribution | -10.18 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
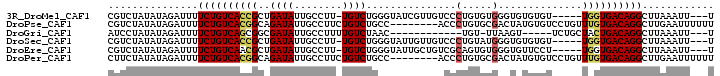
>3R_DroMel_CAF1 6259965 92 + 27905053 CGUCUAUAUAGAUUUUCUGUCACCGCUGAUAUUGCCUU-UGUCUGGGUAUCGUUGUCCCUGUGUGGGUGUGUGU-----UGGUGACAGGCUUAAAUU---U .((((....))))..(((((((((...(((..(((((.-.....)))))..)))..(((.....))).......-----.)))))))))........---. ( -25.30) >DroPse_CAF1 20969 93 + 1 CGUCUAUAUAGAUUUUCUGUCACGGCAGAUAUUGCCUUCUGUCUGCC--------ACCCUGUGCGACUAUGUGUCCUGUUUGUGACAGGCUUGAAUUUUUU .((((....))))..((((((((((((((((........))))))))--------.........(((.....)))......))))))))............ ( -27.10) >DroGri_CAF1 34787 80 + 1 AUCCUAUAUAGAUUUUCUGUCAGCGGCGAUAUUGCCUUUUGUCUAAC------------UGU-UUAAGU-----UCUGCUACUGACAGGCUUAAAUU---U .........((((((((((((((..((((((........)))).(((------------(..-...)))-----)..))..))))))))...)))))---) ( -19.50) >DroSec_CAF1 25221 92 + 1 CGUCUAUAUAGAUUUUCUGUCACCGCUGAUAUUGCCUU-UGUCUGGGUAUUGUUGUCCCUGUAUGGGUGUGUGU-----UGGUGACAGGCUUAAAUU---U .((((....))))..(((((((((...((((.(((((.-.....))))).))))..(((.....))).......-----.)))))))))........---. ( -26.80) >DroEre_CAF1 30940 92 + 1 CGUCUAUAUAGAUUUUCUGUCAACGCUGAUAUUGCCUU-UGUCUGGGUAUUGCUGUCGCAGUGUGGGUGUUCCU-----UGGUGACAGGCUUAAAUU---U .((((....))))..((((((((((((.(((((((((.-.....))))).(((....))))))).)))))....-----...)))))))........---. ( -21.81) >DroPer_CAF1 26029 93 + 1 CUUCUAUAUAGAUUUUCUGUCACGGCAGAUAUUGCCUUCUGUCUGCC--------ACCCUGUGCGACUAUGUGUCCUGUUUGUGACAGGCUUGAAUUUUUU .........((((((((((((((((((((((........))))))))--------.........(((.....)))......))))))))...))))))... ( -27.00) >consensus CGUCUAUAUAGAUUUUCUGUCACCGCUGAUAUUGCCUU_UGUCUGGC________UCCCUGUGUGAGUGUGUGU_____UGGUGACAGGCUUAAAUU___U ...............((((((((((..((((........))))...............(.....)..............))))))))))............ (-10.24 = -10.18 + -0.06)
| Location | 6,259,965 – 6,260,057 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 73.20 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -8.97 |
| Energy contribution | -9.33 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
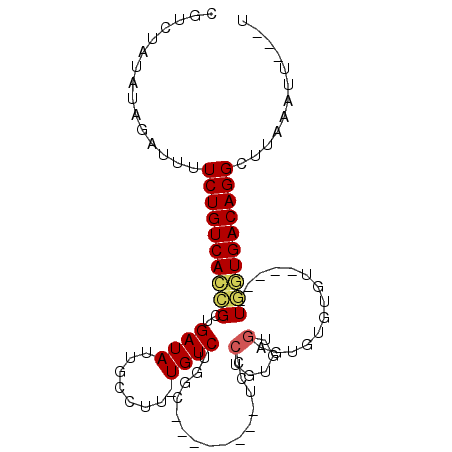
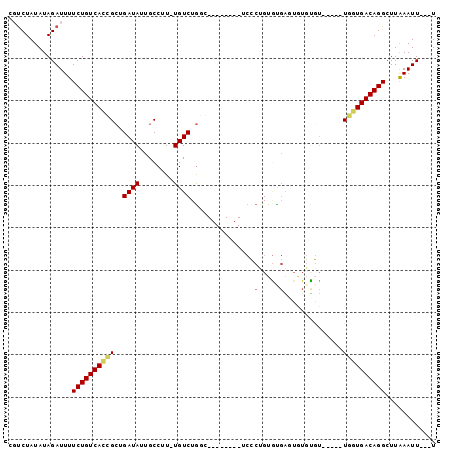
>3R_DroMel_CAF1 6259965 92 - 27905053 A---AAUUUAAGCCUGUCACCA-----ACACACACCCACACAGGGACAACGAUACCCAGACA-AAGGCAAUAUCAGCGGUGACAGAAAAUCUAUAUAGACG .---.........((((((((.-----.......(((.....))).....((((.((.....-..))...))))...))))))))................ ( -21.60) >DroPse_CAF1 20969 93 - 1 AAAAAAUUCAAGCCUGUCACAAACAGGACACAUAGUCGCACAGGGU--------GGCAGACAGAAGGCAAUAUCUGCCGUGACAGAAAAUCUAUAUAGACG .............(((((((..............(((((.....))--------)))........((((.....)))))))))))................ ( -23.10) >DroGri_CAF1 34787 80 - 1 A---AAUUUAAGCCUGUCAGUAGCAGA-----ACUUAA-ACA------------GUUAGACAAAAGGCAAUAUCGCCGCUGACAGAAAAUCUAUAUAGGAU .---.........((((((((.....(-----(((...-..)------------)))........(((......)))))))))))...((((.....)))) ( -18.10) >DroSec_CAF1 25221 92 - 1 A---AAUUUAAGCCUGUCACCA-----ACACACACCCAUACAGGGACAACAAUACCCAGACA-AAGGCAAUAUCAGCGGUGACAGAAAAUCUAUAUAGACG .---.........((((((((.-----.......(((.....))).................-...((.......))))))))))................ ( -20.00) >DroEre_CAF1 30940 92 - 1 A---AAUUUAAGCCUGUCACCA-----AGGAACACCCACACUGCGACAGCAAUACCCAGACA-AAGGCAAUAUCAGCGUUGACAGAAAAUCUAUAUAGACG .---.......(((((((....-----.((.....))....(((....))).......))).-.))))........((((.(.((.....)).)...)))) ( -14.00) >DroPer_CAF1 26029 93 - 1 AAAAAAUUCAAGCCUGUCACAAACAGGACACAUAGUCGCACAGGGU--------GGCAGACAGAAGGCAAUAUCUGCCGUGACAGAAAAUCUAUAUAGAAG .............(((((((..............(((((.....))--------)))........((((.....)))))))))))................ ( -23.10) >consensus A___AAUUUAAGCCUGUCACCA_____ACACACACCCACACAGGGA________CCCAGACA_AAGGCAAUAUCAGCGGUGACAGAAAAUCUAUAUAGACG .............((((((((.......................................(....)((.......))))))))))................ ( -8.97 = -9.33 + 0.36)

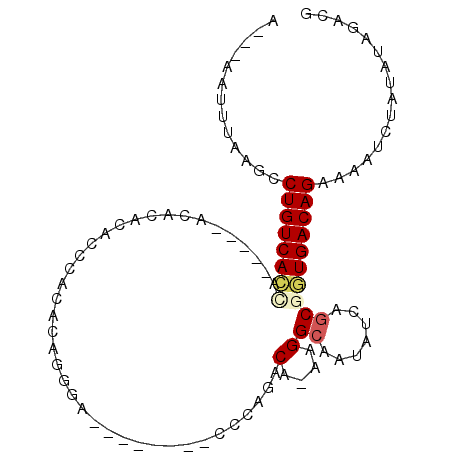
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:28 2006