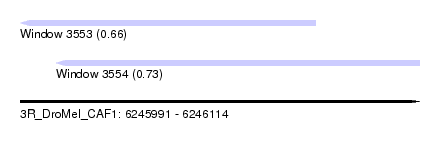
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,245,991 – 6,246,114 |
| Length | 123 |
| Max. P | 0.725114 |

| Location | 6,245,991 – 6,246,082 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.56 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -23.45 |
| Energy contribution | -24.15 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6245991 91 - 27905053 ACACGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGAAUGAAUACCCGGCCGCGAAAUGGCGGCGGGUGAUAAAACACGUUUUAAUAUCAG----------------------------- ....(((((.....)))))..((..(......)..)).(((.((((((.((((......)))))))))).(((((....)))))...))).----------------------------- ( -27.00) >DroVir_CAF1 9177 108 - 1 ACAGGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGAAUGAAUACUCGGCCGCGAAAUGGCGGCGCGUGAUAAAACACGUUUUAAUAUCGA----GGCUCA---CUCCGG-CUCCA---- .((((((((.....)))))...)))(((......(((.(((...(((((((((......)))))(((((......)))))........)))----)..)))---.)))))-)....---- ( -36.30) >DroPse_CAF1 6878 101 - 1 ACACGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGACUGAAUACCCGGCCGCGAAAUGGCGGCGCGUGAUAAAACACGUUUUAAUAUCUG----AGCCCC---C--AG------A---- ....(((((.....)))))..(((((..((..(..((...........(((((......)))))(((((......))))).......))..----)))..)---)--))------)---- ( -31.80) >DroGri_CAF1 14828 116 - 1 ACAGGCACUUGAAAAGUGCAAUCUGGCCGCAAUCGGAAUGAAUACUCGGCCGCGAAAUGGCGGCGCGUGAUAAAACACGUUUUAAUAUCAG----GGCUCAGGACUCUGGACUCUGGACU .((((((((.....)))))...)))(((((..((((..(((....)))..).)))....)))))(((((......))))).......((((----((..(((....)))..))))))... ( -37.30) >DroWil_CAF1 12357 105 - 1 ACACGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGUAUGAAUGCCCGGCCGCGAAAUGGCGGCGCGUGAUAAAACACGUUUUAAUAUCCAGCCAGGCCUC---A--CA------A---- ....(((((.....))))).....((((....(((((((.........(((((......)))))(((((......)))))....))).))))...))))..---.--..------.---- ( -33.30) >DroMoj_CAF1 16248 109 - 1 ACAGGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGAAUGAAUACCCGGCCGCGAAAUGGUGGCGCGUGAUAAAACACGUUUUAAUAUCGG----GGCUCA---CUCUGA-CUCCGG--- .((((((((.....)))))...))).(((...(..((.(((...(((((((((......)))))(((((......)))))........)))----)..)))---.))..)-...)))--- ( -36.40) >consensus ACACGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGAAUGAAUACCCGGCCGCGAAAUGGCGGCGCGUGAUAAAACACGUUUUAAUAUCAG____GGCUCA___C__CG______A____ ....(((((.....))))).....((((....................(((((......)))))(((((......)))))...............))))..................... (-23.45 = -24.15 + 0.70)



| Location | 6,246,002 – 6,246,114 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -21.58 |
| Energy contribution | -21.63 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
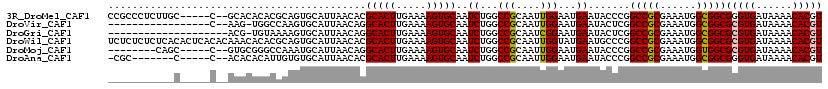
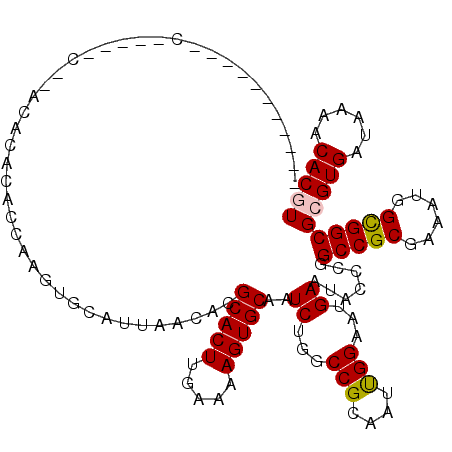
>3R_DroMel_CAF1 6246002 112 - 27905053 CCGCCCUCUUGC-----C--GCACACACGCAGUGCAUUAACACGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGAAUGAAUACCCGGCCGCGAAAUGGCGGCGGGUGAUAAAACACGU .(((((....((-----(--((.((..(((.(((......)))(((((.....))))).....(((((.....((........))))))))))...))))))))))))........... ( -39.50) >DroVir_CAF1 9205 99 - 1 -----------------C--AAG-UGGCCAAGUGCAUUAACAGGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGAAUGAAUACUCGGCCGCGAAAUGGCGGCGCGUGAUAAAACACGU -----------------.--..(-((((((..((((((..(((....)))...))))))...)))))))..................(((((......)))))(((((......))))) ( -35.20) >DroGri_CAF1 14864 98 - 1 --------------------ACG-UGUAAAAGUGCAUUAACAGGCACUUGAAAAGUGCAAUCUGGCCGCAAUCGGAAUGAAUACUCGGCCGCGAAAUGGCGGCGCGUGAUAAAACACGU --------------------.((-.(((.((((((........))))))........((.(((((......))))).))..))).))(((((......)))))(((((......))))) ( -29.90) >DroWil_CAF1 12382 119 - 1 UCUCUCUCUCACACUCACACAAACACACGCAGUGCAUUAACACGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGUAUGAAUGCCCGGCCGCGAAAUGGCGGCGCGUGAUAAAACACGU .........................(((((.(.(((((.....(((((.....)))))......((((....))))...))))).).(((((......))))))))))........... ( -33.30) >DroMoj_CAF1 16277 104 - 1 --------CAGC-----C--GUGCGGGCCAAAUGCAUUAACAGGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGAAUGAAUACCCGGCCGCGAAAUGGUGGCGCGUGAUAAAACACGU --------...(-----(--.(((((.(((..((((((..(((....)))...))))))...))))))))...))............(((((......)))))(((((......))))) ( -32.80) >DroAna_CAF1 18902 104 - 1 -CGC-------C-----C--ACACACAUUGUGUGCAUUAACACGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGAAUGAAUACCCGGCCGCGAAAUGGCGGCGGGUGAUAAAACACGU -.((-------(-----(--((((.....))))).........(((((.....))))).....)))...............((((((.((((......))))))))))........... ( -33.70) >consensus ___________C_____C__ACACACACCAAGUGCAUUAACACGCACUUGAAAAGUGCAAUCUGGCCGCAAUUGGAAUGAAUACCCGGCCGCGAAAUGGCGGCGCGUGAUAAAACACGU ...........................................(((((.....)))))..((...(((....)))...)).......(((((......)))))(((((......))))) (-21.58 = -21.63 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:22 2006