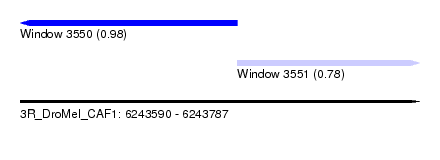
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,243,590 – 6,243,787 |
| Length | 197 |
| Max. P | 0.983913 |

| Location | 6,243,590 – 6,243,697 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.09 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -32.62 |
| Energy contribution | -33.42 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6243590 107 - 27905053 UGGGUCUUCAGUGGGUGUUCCGCACAGAAGGGUGGGGCU---UGGACCAGGGGUUCCAUGUUGACGAAGGUUUUUUGGAGCCGAGGCUCAUGGGGACUUAAAAUCACAUU ((((((((((.((((((((((((........))))))).---.........(((((((....(((....)))...)))))))...))))))))))))))).......... ( -40.10) >DroSec_CAF1 6480 107 - 1 UGGGUCUUCAGUGGGUCUUCCGCACCGAAGGGUGGGGAU---UGGACCUGGGGUUCCCUGUUGACGAAGGUUUUUUGGAGGCGAGGCUUAUGGGGACUUAAAAUCACAUU ((((((((((..(((((((((.((((....)))).))).---.))))))(((.((((((.(.(((....)))....).))).))).))).)))))))))).......... ( -43.90) >DroSim_CAF1 9354 107 - 1 UGGGACUUCAGUGGGUCUUCCGCACCGAAGGGUGGGCAU---UGGACCAGGGGUUCCAUGUUGACGAAGGUUUUUUGGAGGCGAGGCUCAUGGGGACUUAAAAUAACAUU .((((((((....(((((.((.((((....))))))...---.))))).))))))))((((((....((((((((..(((......)))..))))))))....)))))). ( -37.20) >DroEre_CAF1 11134 105 - 1 UGGGUCCUCAGUGGGUCUUCCGC--CGAAGGGUGGGUCU---UUCACCAGGGGUUCCAUGUUGACGAAGGUCUUUUGGAGGCGAGGCUCAUGGGGAUUUAAAAUCACAUU ((((((((((.((((((((((((--(....)))))....---.........(.(((((....(((....)))...))))).))))))))))))))))))).......... ( -43.40) >DroYak_CAF1 9185 108 - 1 UGGGUCUUCAGUGGGUCUUCCGC--CGAAGGGUGGGUCCAUCUCCACCAGGGGUUCCACGUUGACGAAGGUCUUUUGGAGGCGAGGCUCAUGGGGAUUUAAAAUCACAUU ((((((((((.((((((((((((--(....)))))(((...((((....)))).((((....(((....)))...))))))))))))))))))))))))).......... ( -42.70) >consensus UGGGUCUUCAGUGGGUCUUCCGCACCGAAGGGUGGGGCU___UGGACCAGGGGUUCCAUGUUGACGAAGGUUUUUUGGAGGCGAGGCUCAUGGGGACUUAAAAUCACAUU ((((((((((.((((((((((.((((....))))((..........)).))(.(((((....(((....)))...))))).))))))))))))))))))).......... (-32.62 = -33.42 + 0.80)


| Location | 6,243,697 – 6,243,787 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -16.85 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6243697 90 + 27905053 CCGA--ACCC----ACCAACGACACUUUGAGGCCCCAUAACGCGGGCACACGCAUAAUGAUCCUUUGAAUGGCUCAAUGCGUGGCUCCGUUCCAAC ..((--((..----.................(((((.....).)))).(((((((..(((.((.......)).)))))))))).....)))).... ( -24.30) >DroPse_CAF1 4789 85 + 1 AUCC--A-CC----GCCAACGACACUUUGAGGCCCCAUAACGCGGGCACACGCAUAAUGAUCCUUUGAAUGGCUCAAUGCGUGGCC----UCCACC ....--.-..----..............((((((((.......)))..(((((((..(((.((.......)).)))))))))))))----)).... ( -25.20) >DroGri_CAF1 11934 76 + 1 UUCA--A-CC----AAAAACGACACUUUUAGGCGCCAUAACGUGGGCGCACGCAUAAUGAUCCCCCGAAUGGCUCAAUGCGUA------------- ....--.-..----.................(((((........)))))((((((..(((.((.......)).))))))))).------------- ( -19.50) >DroYak_CAF1 9293 92 + 1 CCGAAAACCC----ACCAACGACACUUUGAGGCCCCAUAACGCGGGCACACGCAUAAUGAUCCUUUGAAUGGCUCAAUGCGUGGAACCGCUCCAGC ..........----..............((((((((.....).)))).(((((((..(((.((.......)).))))))))))......))).... ( -23.40) >DroAna_CAF1 16779 92 + 1 UUCG--ACCCCCAUCCCGACGACACUC-GAGA-CCCAUAACGCGGGCGCACGCAUAAUGAUCCUGUGAAUGGCUCAAUGCGUGUUCUGGCUCCACC ....--.......((.(((......))-).))-........((.((.((((((((..(((.((.......)).))))))))))).)).))...... ( -23.70) >DroPer_CAF1 4737 85 + 1 AUCC--A-CC----GCCAACGACACUUUGAGGCCCCAUAACGCGGGCACACGCAUAAUGAUCCUUUGAAUGGCUCAAUGCGUGGCC----UCCACC ....--.-..----..............((((((((.......)))..(((((((..(((.((.......)).)))))))))))))----)).... ( -25.20) >consensus AUCA__A_CC____ACCAACGACACUUUGAGGCCCCAUAACGCGGGCACACGCAUAAUGAUCCUUUGAAUGGCUCAAUGCGUGGCC____UCCACC ...............................(((((.....).)))).(((((((..(((.((.......)).))))))))))............. (-16.85 = -17.52 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:20 2006