
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,233,003 – 6,233,156 |
| Length | 153 |
| Max. P | 0.984677 |

| Location | 6,233,003 – 6,233,123 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -22.40 |
| Energy contribution | -24.58 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6233003 120 + 27905053 AACAAAAAUCUAUGUUGGUUCGAUACUAUCAACAACAAUGCCAGUAGCAUUAGUUAUCGAUAAGAACAAAAUCAAGAAUCGUAAUUGAACUAUCGAUAGAUAGAGCUUCAAGGGCGGUUU .....(((((..(((((((........))))))).....(((...(((.(((((((.((((..((......))....))))))))))).(((((....))))).))).....)))))))) ( -26.50) >DroSec_CAF1 6049 120 + 1 ACCAAAAAUCUAUGUUGAUUCGAUACUAUCAACAACAAUGUCAGUAGCAUUAGUUAUCGAUAAGAACCAAAUCAAGAAUAAUAAUUGACUUAUCGAUAGAUAGAGCUGCAAGGGCGGUUU (((.........(((((((........))))))).....(((.(((((.(((.(((((((((((.......((((.........))))))))))))))).))).)))))...)))))).. ( -31.91) >DroSim_CAF1 8332 120 + 1 AACAAAAAUCUAUGUUGAUUCGAUACUAUCAACAACAAUGUCAGUAGCAUGAGUUAUCGAUAAGAACAAAAUCAAGAAUAAUAAUUGACUUAUCGAUAUAUAGAGCUACAAGGGCGGUUU .....(((((..(((((((........))))))).....(((.(((((......((((((((((.......((((.........))))))))))))))......)))))...)))))))) ( -28.41) >DroYak_CAF1 6201 120 + 1 AACAAAAUUCUAUGUUGAUUCGAUACUAUCAACAACAACGUGAGUAGCUGUAGUUAUCGAUUGAACUACAAUCAAUAAUUGAAGUUGACUUAUCGAUAUGAAGCACUACUAGGGCGGUUU ............(((((((........)))))))....(((.(((((.(((...((((((((.((((.((((.....)))).)))).)...)))))))....))))))))...))).... ( -27.30) >consensus AACAAAAAUCUAUGUUGAUUCGAUACUAUCAACAACAAUGUCAGUAGCAUUAGUUAUCGAUAAGAACAAAAUCAAGAAUAAUAAUUGACUUAUCGAUAGAUAGAGCUACAAGGGCGGUUU ............(((((((........)))))))....((((.(((((.(((.(((((((((((.......((((.........))))))))))))))).))).)))))...)))).... (-22.40 = -24.58 + 2.19)



| Location | 6,233,003 – 6,233,123 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.86 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6233003 120 - 27905053 AAACCGCCCUUGAAGCUCUAUCUAUCGAUAGUUCAAUUACGAUUCUUGAUUUUGUUCUUAUCGAUAACUAAUGCUACUGGCAUUGUUGUUGAUAGUAUCGAACCAACAUAGAUUUUUGUU .........(((((...(((((....))))))))))..((((.(((.......(((((((((((((((.(((((.....))))))))))))))))....))))......)))...)))). ( -25.72) >DroSec_CAF1 6049 120 - 1 AAACCGCCCUUGCAGCUCUAUCUAUCGAUAAGUCAAUUAUUAUUCUUGAUUUGGUUCUUAUCGAUAACUAAUGCUACUGACAUUGUUGUUGAUAGUAUCGAAUCAACAUAGAUUUUUGGU ..(((((....))......((((((.(((((.....)))))....((((((((((..(((((((((((.(((((....).))))))))))))))).)))))))))).))))))....))) ( -28.60) >DroSim_CAF1 8332 120 - 1 AAACCGCCCUUGUAGCUCUAUAUAUCGAUAAGUCAAUUAUUAUUCUUGAUUUUGUUCUUAUCGAUAACUCAUGCUACUGACAUUGUUGUUGAUAGUAUCGAAUCAACAUAGAUUUUUGUU ...........(((((......((((((((((.(((..((((....)))).)))..))))))))))......))))).((((....(((((((........)))))))........)))) ( -26.80) >DroYak_CAF1 6201 120 - 1 AAACCGCCCUAGUAGUGCUUCAUAUCGAUAAGUCAACUUCAAUUAUUGAUUGUAGUUCAAUCGAUAACUACAGCUACUCACGUUGUUGUUGAUAGUAUCGAAUCAACAUAGAAUUUUGUU .....((.(.....).))......((((((.((((((.....((((((((((.....))))))))))..(((((.......))))).))))))..))))))...((((........)))) ( -25.10) >consensus AAACCGCCCUUGUAGCUCUAUAUAUCGAUAAGUCAAUUACUAUUCUUGAUUUUGUUCUUAUCGAUAACUAAUGCUACUGACAUUGUUGUUGAUAGUAUCGAAUCAACAUAGAUUUUUGUU ...........(((((......((((((((((((((.........)))).......))))))))))......))))).........(((((((........)))))))............ (-16.80 = -17.86 + 1.06)

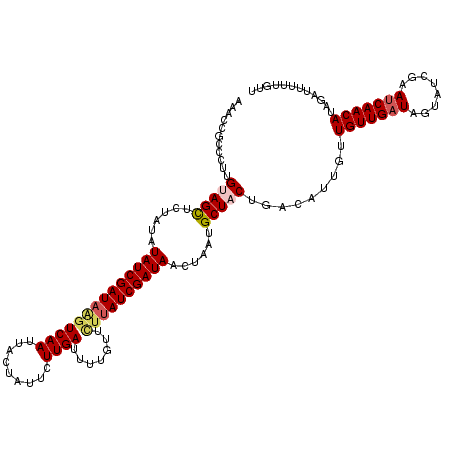

| Location | 6,233,043 – 6,233,156 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -16.11 |
| Energy contribution | -17.86 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6233043 113 + 27905053 CCAGUAGCAUUAGUUAUCGAUAAGAACAAAAUCAAGAAUCGUAAUUGAACUAUCGAUAGAUAGAGCUUCAAGGGCGGUUUGUUCAAAUGCUAGUUUCUGUAGCUAUGAAUUAA--- ...(((((...............((((((........(((((..((((((((((....)))))...)))))..)))))))))))...(((........)))))))).......--- ( -24.60) >DroSec_CAF1 6089 111 + 1 UCAGUAGCAUUAGUUAUCGAUAAGAACCAAAUCAAGAAUAAUAAUUGACUUAUCGAUAGAUAGAGCUGCAAGGGCGGUUUGUUCAAAUGCUACUUUCUGUAGAUAUGAAUU----- ..(((((((((...((((((((((.......((((.........))))))))))))))((..(((((((....)))))))..)).))))))))).................----- ( -30.51) >DroSim_CAF1 8372 111 + 1 UCAGUAGCAUGAGUUAUCGAUAAGAACAAAAUCAAGAAUAAUAAUUGACUUAUCGAUAUAUAGAGCUACAAGGGCGGUUUGUUCAAAUGCUACUUUCUGUAGAUAUGAAUU----- ...(((((......((((((((((.......((((.........))))))))))))))......)))))...........(((((.((.((((.....)))))).))))).----- ( -24.81) >DroYak_CAF1 6241 116 + 1 UGAGUAGCUGUAGUUAUCGAUUGAACUACAAUCAAUAAUUGAAGUUGACUUAUCGAUAUGAAGCACUACUAGGGCGGUUUGUUUAAAUAUUAGUUUCUGUAGGUAUGAACUGCUAU ..(((((.(((...((((((((.((((.((((.....)))).)))).)...)))))))....))))))))..(((((((..(.....(((........)))...)..))))))).. ( -27.80) >consensus UCAGUAGCAUUAGUUAUCGAUAAGAACAAAAUCAAGAAUAAUAAUUGACUUAUCGAUAGAUAGAGCUACAAGGGCGGUUUGUUCAAAUGCUACUUUCUGUAGAUAUGAAUU_____ ...(((((.(((.(((((((((((.......((((.........))))))))))))))).))).)))))...........(((((..(((........)))....)))))...... (-16.11 = -17.86 + 1.75)



| Location | 6,233,043 – 6,233,156 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -11.92 |
| Energy contribution | -12.73 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6233043 113 - 27905053 ---UUAAUUCAUAGCUACAGAAACUAGCAUUUGAACAAACCGCCCUUGAAGCUCUAUCUAUCGAUAGUUCAAUUACGAUUCUUGAUUUUGUUCUUAUCGAUAACUAAUGCUACUGG ---..............(((....((((((((((((.....((.......))..((((....)))))))))....((((....((......))..))))......)))))))))). ( -17.70) >DroSec_CAF1 6089 111 - 1 -----AAUUCAUAUCUACAGAAAGUAGCAUUUGAACAAACCGCCCUUGCAGCUCUAUCUAUCGAUAAGUCAAUUAUUAUUCUUGAUUUGGUUCUUAUCGAUAACUAAUGCUACUGA -----.................(((((((((.(........((....)).........((((((((((......((((....))))......)))))))))).).))))))))).. ( -24.50) >DroSim_CAF1 8372 111 - 1 -----AAUUCAUAUCUACAGAAAGUAGCAUUUGAACAAACCGCCCUUGUAGCUCUAUAUAUCGAUAAGUCAAUUAUUAUUCUUGAUUUUGUUCUUAUCGAUAACUCAUGCUACUGA -----..((((.((((((.....)))).)).))))............(((((......((((((((((.(((..((((....)))).)))..))))))))))......)))))... ( -25.90) >DroYak_CAF1 6241 116 - 1 AUAGCAGUUCAUACCUACAGAAACUAAUAUUUAAACAAACCGCCCUAGUAGUGCUUCAUAUCGAUAAGUCAACUUCAAUUAUUGAUUGUAGUUCAAUCGAUAACUACAGCUACUCA .((((((((..(......)..))))......................(((((......(((((((.....((((.(((((...))))).))))..)))))))))))).)))).... ( -17.10) >consensus _____AAUUCAUAUCUACAGAAACUAGCAUUUGAACAAACCGCCCUUGUAGCUCUAUAUAUCGAUAAGUCAAUUACUAUUCUUGAUUUUGUUCUUAUCGAUAACUAAUGCUACUGA ...............................................(((((......((((((((((((((.........)))).......))))))))))......)))))... (-11.92 = -12.73 + 0.81)

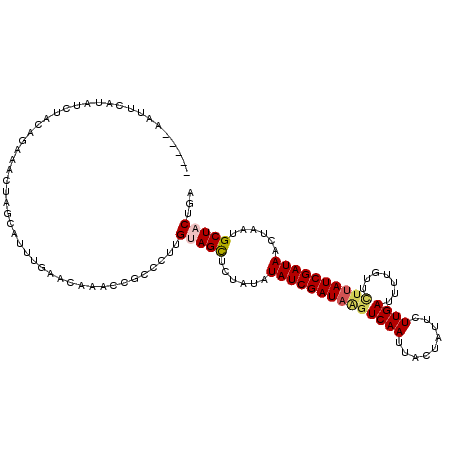
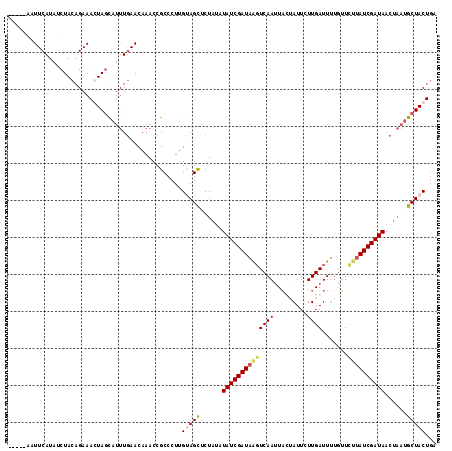
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:10 2006